CLARITY and 3D imaging with high resolution and deep scanning of innervation in the pig colon by using SP8 DIVE fully tunable spectral multiphoton microscope
Deep scanning of innervation in the pig colon by using SP8 DIVE multiphoton microscope
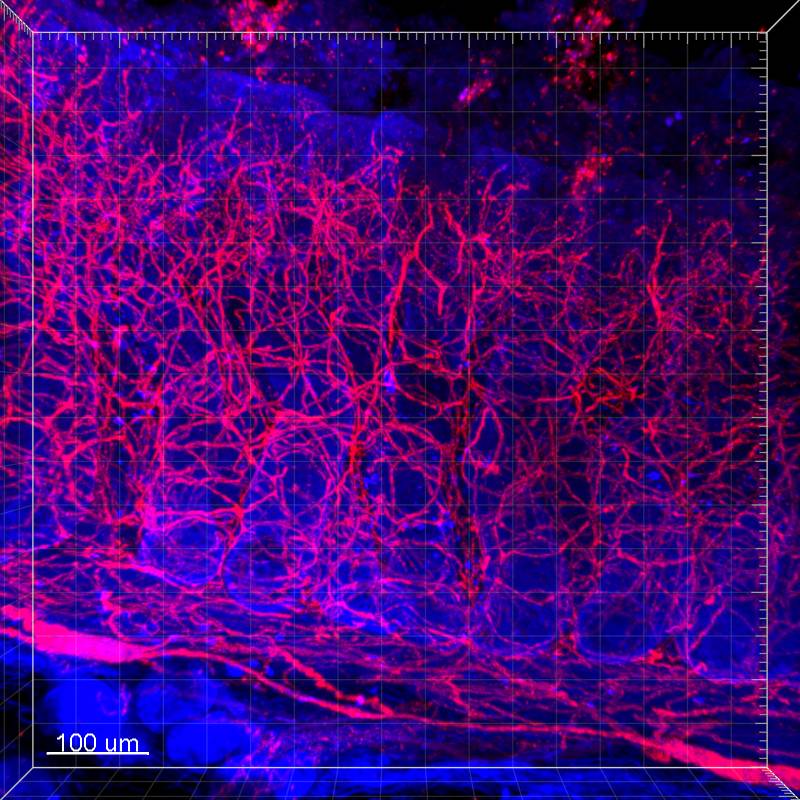
Dataset Overview
Study Purpose: To demonstrate the innervation of the pig proximal colon mucosa in 3D structure with high resolution and deep scanning by using SP8 DIVE multiphoton microscope, the latest multiphoton microscope with fully spectrally tunable detection.
Data Collection: The pig proximal colon samples were cleared by a modified passive CLARITY protocol we developed in the first SPARC funding year. Immunofluorescent methods were used to label the nerve fibers in the mucosa by using antibodies against PGP9.5, Neurofilement (NF), and vasoactive intestinal polypeptide (VIP). Images were acquired with Leica TCS SP8 DIVE multiphoton microscope (Leica Microsystem). The Z-axis intervals were 1 µm with depths between 500 – 1000 µm. The images were collected and processed for 3D reconstruction and video generation using Leica Application Suite software (LAS X 3.0, Leica Microsystem) and Imaris 9.2 for Neuroscientists (Bitplane Inc., Concord, MA). The projections of these fine fibers from inner submucosal neurons and sprouting into mucosa with multiple branches were also clearly displayed by videos in a 360° horizontal or vertical rotation.
Primary Conclusion: The dataset of 3D images and videos of innervation in the pig colonic mucosa obtained by using CLARITY/SP8 DIVE multiphoton microscope with high resolution and deep scanning provides fundamental information to distinguish the different classes of nerve fibers, visualize their projections to different layers and compare their differences among the segments, which are not readily revealed by conventional confocal microscope and rarely reported before.
Curator's Notes
Experimental Design: Cleared samples were cut into 0.5–1 mm vertical sections using a vibratome after hydrogel polymerization, followed by washing with clearing solution as described in the CLARITY protocol for 1-2 weeks. Samples were immersed in 10% normal donkey serum in 0.3% triton/PBS at room temperature for one day and then incubated with primary antibodies: rabbit anti-protein gene product 9.5 (PGP9.5, 1:1000, RRID:AB_10891773), rabbit anti-vasoactive intestinal peptide (VIP, 1:1000, RRID:AB_2890602), chicken anti-neurofilament 200 (NF, 1:2000, RRID:AB_11212161) at RT for two days. After being washed with PBS at RT for one day, samples were incubated in Alexa 488-conjugated anti-rabbit IgG (for PGP9.5), Alexa 555-conjugated anti-rabbit IgG (for VIP) or Alexa 555-conjugated anti-chicken IgG (1:400, Jackson Immuno Research) at RT with shaking for one day and then washed with PBS at RT for one day. After counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 1µg/ml, Sigma), samples were then immersed in a custom-made refractive index matching solution (RIMS) containing 88% Histodenz (Sigma-Aldrich) in 0.02 M phosphate buffer with 0.01% sodium azide (pH 7.5) with a refractive index of 1.46 at 4°C (8) until transparent (1 day for 0.5–1 mm vertical sections, 2-3 days for 1×1 cm SMP a with iSpacers (SunJin Lab, Hsinchu City, Taiwan) which are made from different thickness adhesive tape. Images were acquired with Leica TCS SP8 DIVE multiphoton microscope (Leica Microsystem). The Z-axis intervals were 1 µm with depths between 500 – 1000 µm. The images were collected and processed for 3D reconstruction and video generation using Leica Application Suite software (LAS X 3.0, Leica Microsystem) and Imaris 9.2 for Neuroscientists (Bitplane Inc., Concord, MA).
Completeness: this dataset is complete.
Subjects & Samples: Male (n=2) and female (n=1) adult yucatan pigs were used in this study.
Primary vs derivative data: Primary data is organized by subject ID. Each sample subfolder contains videos showing the deep scanning of porcine colonic mucosa innervation from inner submucosa plexus (.mp4), images with high magnification showing the porcine colonic mucosa innervation from inner submucosa plexus (.tif) and files with a 3D reconstruction of innervation (.ims). The derivative folder contains converted primary microscopy images.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Yuan, P.-Q., & Taché, Y. (2019). Tache_Yuan_OT2OD024899_CLARITYAnd3DImagingOfColonicENSintheMouseAndPig_1_2019-Pig_Protocol v1. https://doi.org/10.17504/protocols.io.4r9gv96
