Topographical mapping of sympathetic postganglionic innervation of the mouse heart
Anatomical mapping of sympathetic efferent projection to the mouse heart
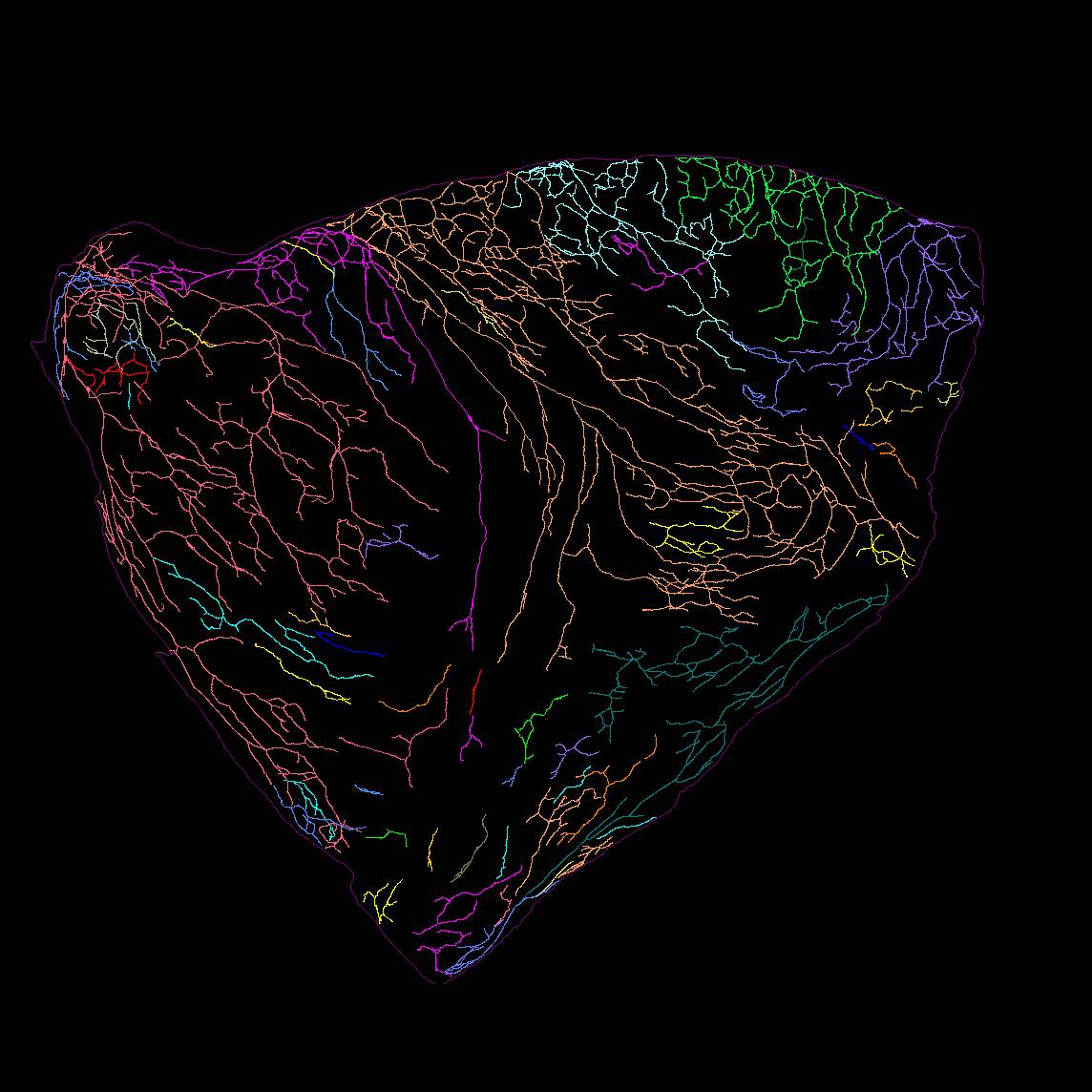
Dataset Overview
Study Purpose: To map the topographical distribution of the sympathetic postganglionic innervation in whole atria of mice. Our work will provide a comprehensive topographical map of the catecholaminergic axon distribution and morphology innervation of the whole heart at single cell/axon/synapse scale that will provide a cardiac sympathetic-brain atlas.
Data Collection: State-of-the-art imaging (Confocal and Fluorescence Microscopy), Neurolucida 360.
Primary Conclusion: We found that (1) 4–5 major extrinsic tyrosine hydroxylase (TH)-immunoreactive (IR) nerve bundles entered the atria at the superior vena cava, right atrium (RA), left precaval vein, and the root of the pulmonary veins (PVs) in the left atrium (LA). Although these bundles projected to different areas of the atria, their projection fields partially overlapped. (2) TH-IR axon and terminal density varied considerably between different sites of the atria, with the greatest density of innervation near the sinoatrial node region (P < 0.05, n = 6). (3) TH-IR axons also innervated blood vessels and adipocytes. (4) Many principal neurons in intrinsic cardiac ganglia and small intensely fluorescent cells were also strongly TH-IR.
Curator's Notes
Experimental Design: In this study, we used Tyrosine hydroxylase (TH) as a marker for sympathetic axons. We applied a combination of state-of-the-art techniques, including confocal microscopy, Zeiss Imager microscopy, flat-mount tissue processing, immunohistochemistry, Neurolucida Tracing, and integration of the tracing data onto a 3D Heart Scaffold.
Completeness: This dataset is complete.
Subjects & Samples: 8 male wild-type mice (RRID:IMSR_JAX000664) were used in this study.
Primary vs derivative data: Data in the primary folder are organized by subject ID, then by sample ID. The primary folder contains either confocal images in a .tif format or Neurolucida 360 Tracing files .xml. The primary images in the derivative folder were converted with 20:1 compression to JPEG2000 (.jp2) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal. The primary images were also converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience. Microscopy image metadata is included in the file header of all .jp2 and .tif in the derivative folder.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Topographical mapping of sympathetic postganglionic innervation of mouse heart v2. (2023). https://doi.org/10.17504/protocols.io.n92ldzbmxv5b/v2
Mapping CGRP-IR innervation of male mice stomach with Neurolucida 360 v1. (2021). https://doi.org/10.17504/protocols.io.bygmptu6
