CLARITY and three-dimensional (3D) imaging of the mouse and porcine colonic innervation
Regional ENS 3D structure/phenotype, axonal projections and connectivity in the mouse and pig colon.
Dataset Overview
Study Purpose: Little information is available about 3D anatomical and phenotypical maps of the colonic enteric nerve system (ENS) that are indispensable for understanding the structure-function relationships. CLARITY is a newly developed clearing technique removing lipids while keeping proteins in the tissue. However, its application has been largely restricted to the mouse brain. We aim to establish CLARITY protocols applicable for clearing the mouse and porcine colon and demonstrate 3D imaging and phenotyping of colonic innervation, especially in the porcine whose colonic ENS resembles the human unlike rodents.
Data Collected: Four male adult C57BL/6J mice and 1 infant female, 1 adult male, 1 adult female Yucatan minipigs were used for CLARITY. The mouse proximal and distal colon (pC and dC), porcine pC, transverse (tC) and dC were dissected. After CLARITY procedures, IHC and multiple labeling of PGP9.5, Hu C/D, neurofilament heavy chain, S100β, peripheral choline acetyltransferase and neuronal nitric oxide synthase were performed. Images acquired with ZEISS LSM710 confocal and SP8 DIVE multiphoton microscope were reconstructed into 3D images and videos using Imaris 9.1 for neuroscientists.
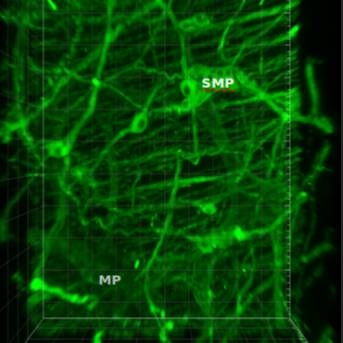
Primary Conclusion: Colonic samples of both mice and pigs were successfully cleared which enable immunohistochemistry (IHC) and maintenance of fluorophores during imaging. 3D analysis revealed in mice: (i) Segmental differences in innervation (pC>dC); (ii) Connectivity between submucosal and myenteric plexus (SP and MP); (iii) Proximity of enteric neurons and enteric glial cells in SP and MP. In porcine: (i) Connectivity between inner SP (ISP) and outer SP (OSP) or OSP and MP; (ii) Rich innervation of the mucosa primarily from ISP; (iii) Distinct patterns of intrinsic cholinergic innervation with ISP>OSP. We have successfully developed CLARITY/IHC protocols applicable for 3D imaging and phenotyping of colonic innervation in the mouse and porcine colon. The information from 3D images and videos, not readily revealed in 2D, provides fundamental information to distinguish the different classes of enteric neurons, visualize their projections to different layers and compare their differences among the segments.
Curator's Notes
Experimental design: Adult male mice and adult minipigs were used. Several tissues were dissected from each animal, Clarity procedures were performed and images were acquired from each sample.
Completeness: This dataset is part of a larger study, called "3D imaging, colon, innervation, enteric nerve system, phenotyping". Related datasets are available.
Subjects & Samples: Juvenile male C57Bl/6J mice, and three male and female minipigs were used
Primary vs derivative data: The raw data is formatted as lsm, which is a Zeiss specific format, but it can be opened using Imaris software. The derived folder contains the same files (also called lsm), but these have been transformed to JPEG 2000 format.Image data (.jpx) was derived from primary images (.czi). Images were converted with 20:1 compression to .jpx by MBF Bioscience for web streaming and visualization on the SPARC Data Portal. Segmentation data (.xml) was derived from Imaris (.ims) marker files in primary folder. The process of converting Imaris (.ims) marker files was performed by MBF Bioscience with Neurolucida 360 (SCR_016788) and converter from MBF Bioscience.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Yuan, P.-Q., Wang, L., & Taché, Y. (2019). Tache_Yuan_OT2OD024899_CLARITYAnd3DImagingOfColonicENSintheMouseAndPig_1_2019-Mouse_Protocol (Annotation Copy) v1. https://doi.org/10.17504/protocols.io.4sagwae
