Mouse genetic models to manipulate enterochromaffin cell activity
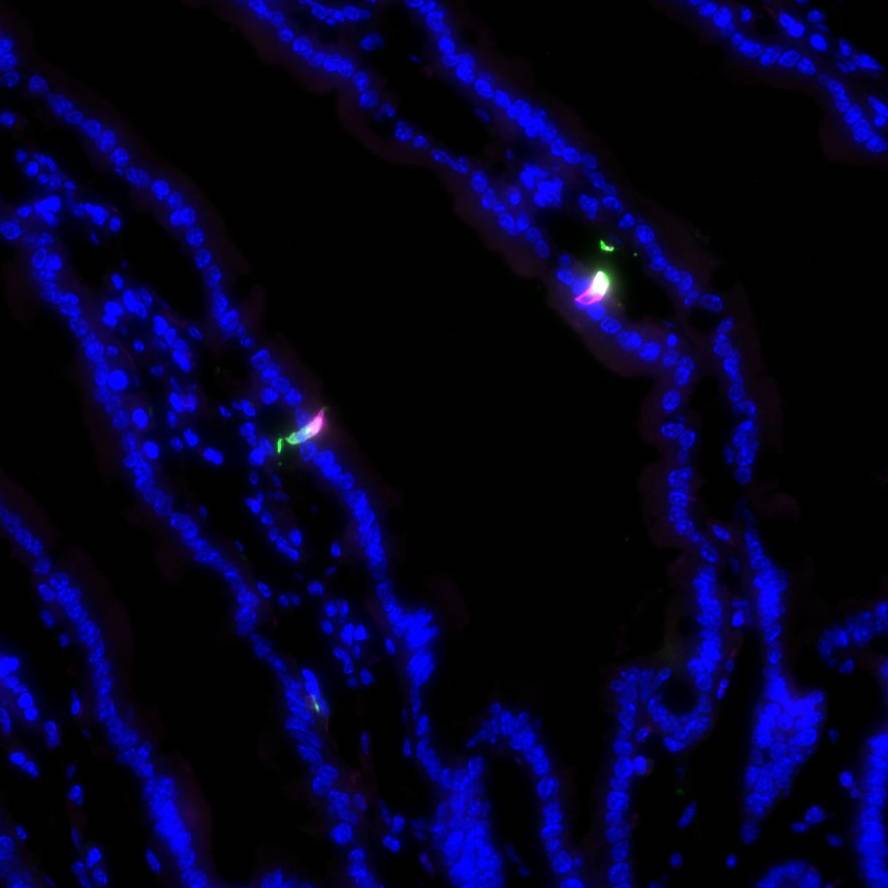
Immunohistochemical validation of mouse genetic strategies used to specifically activate and silence enterochromaffin cells in the intestine
Dataset Overview
Study Purpose: The purpose of this study was to determine if the transgenic mouse lines Pet1Flp;Tac1Cre;RC::PFTox, Pet1Flp;Vil1Cre;RC::PFTox, and Pet1Flp;Tac1Cre;RC::FL-hM3Dq and effectively restrict expression of the effector alleles to enterochromaffin cells.
Data Collection: Confocal imaging was performed on Nikon Ti2 microscope with Crest LFOV spinning disk and Nikon Ti microscope with Yokagawa CSU22 spinning disk. Images were assembled in ImageJ.
Primary Conclusion: The conclusion of this study was that effector allele expression is restricted to enterochromaffin cells.
Curator's Notes
Experimental Design: Mice were euthanized, the small and large intestines collected, and fixed frozen sections of 5 μm and 20μm were prepared. The microscope mounted tissue sections were stained using immunohistochemistry protocols. Antibodies against serotonin (RRID:AB_572263 and RRID:AB_1142794), GFP (RRID:AB_2307313), and mCherry (RRID:AB_10013483) were used. Confocal imaging was performed on a Nikon Ti2 microscope with Crest LFOV spinning disk and Nikon Ti microscope with Yokagawa CSU22 spinning disk. Images were assembled in ImageJ.
Completeness: This dataset is a part of the larger study: Mouse genetic models to manipulate enterochromaffin cell activity.
Subjects & Samples: Male (n=3) adult transgenic mice were used in this study.
Primary vs derivative data: Primary data is organized by the subject ID and then sample ID folders. Each folder contains raw confocal images as .nd2 files. Derivative image data (JPEG2000 and OME-TIFF) was derived from primary images (.ims). Primary images were converted with 20:1 compression to JPEG2000 (.jpx) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Bayrer, J. R., Castro, J., Venkataraman, A., Touhara, K. K., Rossen, N. D., Morrie, R. D., Hendry, A., Madden, J., Braverman, K. N., Schober, G., Brizuela, M., Silva, C. B., Ingraham, H. A., Brierley, S. M., & Julius, D. (2022). Gut Enterochromaffin Cells are Critical Drivers of Visceral Pain and Anxiety. https://doi.org/10.1101/2022.04.04.486775
Is Supplemented by
D Rossen, N. (2022). Preparation and Immunohistochemistry of Mouse Small Intestine and Colon v1. https://doi.org/10.17504/protocols.io.j8nlkw575l5r/v1
