RNA sequencing analysis of transcriptomic responses to vagal nerve stimulation in myenteric ganglia of porcine colon
RNA-seq analysis of porcine colonic ENS responses to VNS
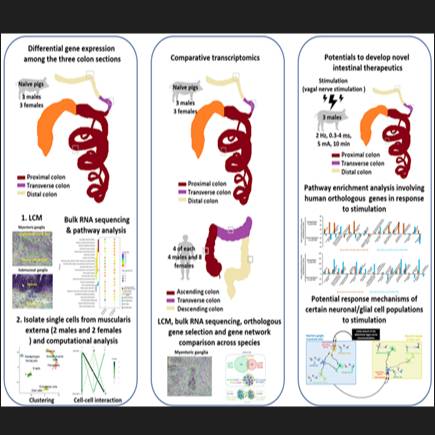
Dataset Overview
Study Purpose: The porcine is a relevant experimental model due to its similarity to the human in nutrition, physiology and metabolic process, particularly in the enteric nervous system (ENS). Thus, the porcine model has untapped potential for a greater understanding of the underlying colonic functions of human in health and diseases. Recent studies in mice have demonstrated heterogeneity in neuronal identity, morphology, projection orientation and synaptic complexity within the ENS located in different colonic segments in line with the diversity of colonic motility patterns. The region-dependent molecular characterization of porcine ENS has been little investigated so far. In addition, colonic functions are modulated through parasympathetic innervation, responsible for regulating colonic secretion and motility. We have recently reported that electrical vagal nerve stimulation (VNS) triggered pan-colonic contractions in the porcine. However, it is unknown how such VNS influences colonic transcriptional programs and whether the impacts of VNS are region-specific. Here, we use laser-capture microdissection (LCM) coupled with bulk RNA sequencing analysis to profile the transcriptomes of myenteric ganglia (MG) from porcine proximal and distal colon (p-pC, p-dC) and to evaluate discrepancy of the region-specific gene programs in MG after electrical stimulation of the celiac branch of the abdominal vagus nerve in anesthetized porcine.
Data Collected: This dataset contains results of RNA-seq analysis of porcine colonic ENS responses to VNS of myenteric ganglia of porcine colon.
Primary Conclusion: (i) When comparing p-pC-MG and p-dC-MG in porcine without VNS, there were 271 upregulated and 212 downregulated genes in p-pC-MG, while 390 upregulated and 265 downregulated genes existed in p-pC-MG in comparison of p-pC-MG and p-dC-MG in porcine with VNS; (ii) Based on comparison of p-pC-MG and p-dC-MG, VNS promoted enrichment of most of the WikiPathways in p-pC-MG, including gap junctions, pro- and anti-inflammatory signaling, chemokine signaling pathway, neurotransmitter uptake and metabolism in glial cells, NO/cGMP/PKG mediated neuroprotection, dopaminergic neurogenesis, nicotine activity on dopaminergic neurons, oncostatin M signaling pathway and TGF-beta receptor signaling, and reduced enrichment of human orthologous risk genes for intestinal and extra-intestinal diseases, synaptic vesicle pathway and neurotransmitter release cycle. By contrast, effects of VNS on p-dC-MG were reflected by improved enrichment of smooth muscle contraction, GABA receptor signaling and glutamate binding, activation of AMPA receptors and synaptic plasticity. In addition, the comparison between p-pC-MG and p-pC-ISG showed that VNS decreased the enrichment scores of most of the WikiPathways in p-pC-MG, such as gap junctions, anti-inflammatory signaling, smooth muscle contraction, chemokine signaling pathway, glutamate binding, activation of AMPA receptors and synaptic plasticity and TGF-beta receptor signaling, while increasing the enrichment scores of the WikiPathways, including pro-inflammatory signaling, acetylcholine binding and downstream events, GABA receptor signaling, neurotransmitter release cycle and synaptic vesicle pathway. The alterations were further explained by the functional similarity between gene products, which was measured by semantic similarities between the annotated BPs of each gene, resulting in similarity matrix. High average similarity scores in the WikiPathways of interest indicate strong functional similarities. Gap junctions and neurotransmitter release cycle had the highest similarity scores. High scores existed when comparing acetylcholine binding and downstream events or neurotransmitter release cycle with pro-inflammatory unlike anti-inflammatory signaling. Apart from functional similarities, the BPs comprised in the WikiPathways were interactive. Pearson correlation analysis revealed a significant positive correlation among gap junctions, smooth muscle contraction and anti-inflammatory signaling and between acetylcholine binding and downstream events and neurotransmitter release cycle. Finally, VNS exerted its effects on p-pC-MG by reducing the enrichment of human orthologous risk genes for intestinal and extra-intestinal diseases and on p-pC-ISG by decreasing the activation of angiotensin pathway, respectively.
Curator's Notes
Experimental Design: Six castrated male Yucatan minipigs were fasted for 12 h and anesthetized. Three out of the six male animals underwent electrical stimulation of the celiac branch of the abdominal vagus nerve (2 Hz, 0.3-4 ms, 5 mA, 10 min) using pulse train. The colonic specimens with full thickness were removed from p-pC and p-dC. All samples were embedded in OCT, snap-frozen in dry ice and stored at -80°C for LCM procedures. A range of 25-40 ganglia/subject were harvested from MG respectively of p-pC and p-dC (3 without VNS and 3 with VSN) using LMD-6000 Laser Micro-dissection System. Total RNA was extracted for construction of cDNA libraries and sequenced on an Illumina HiSeq 3000 sequencer as 50 base pair single-end reads. All of the reads were then aligned to the Sus_scrofa genome using STAR. The Sus_scrofa genome index was created using Sus_scrofa genome file (Sscrofa11.1.fa) and annotation file (Sus_scrofa.Sscrofa11.1.95.gtf), which were downloaded at https://uswest.ensembl.org /Sus_scrofa/Info/Index. The fastq files were then aligned against the genome assembly using STAR, followed by assessment for the total number of aligned reads and total number of uniquely aligned reads to evaluate sequencing performance, with parameters --runMode alignReads --outSAMtype BAM SortedByCoordinate Unsorted --outFilterMultimapNmax 1 --quantMode GeneCounts --twopassMode Basic --sjdbOverhang 50.
Completeness: this dataset is complete.
Subjects & Samples: This study used six adult male Yucatan pigs.
Primary vs derivative data: Primary data folder is organized by the subject ID and then the sample ID. Each sample subfolder contains .fastq.gz raw 50 base pair single end reads generated from Hiseq3000. The derivative data folder contains a spreadsheet with summative gene counts.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Li, T., Morselli, M., Su, T., Mulugeta, M., Larauche, M., Pellegrini, M., Taché, Y., & Yuan, P.-Q. (2022). Comparative transcriptomics reveal highly conserved regional programs between porcine and human colonic enteric nervous system. https://doi.org/10.1101/2022.02.24.480770
Is Supplemented by
Li, T. (2022). RNA sequencing analysis of transcriptomic responses to vagal nerve stimulation in myenteric ganglia of porcine colon v1. https://doi.org/10.17504/protocols.io.eq2lynmeqvx9/v1
