Single cell RNA sequencing of retrogradely labeled mouse stellate ganglion neurons
Datasets from single cell sequencing of murine stellate ganglion neurons retrogradely labeled with cholera toxin B injections into the brown adipose tissue or forelimb
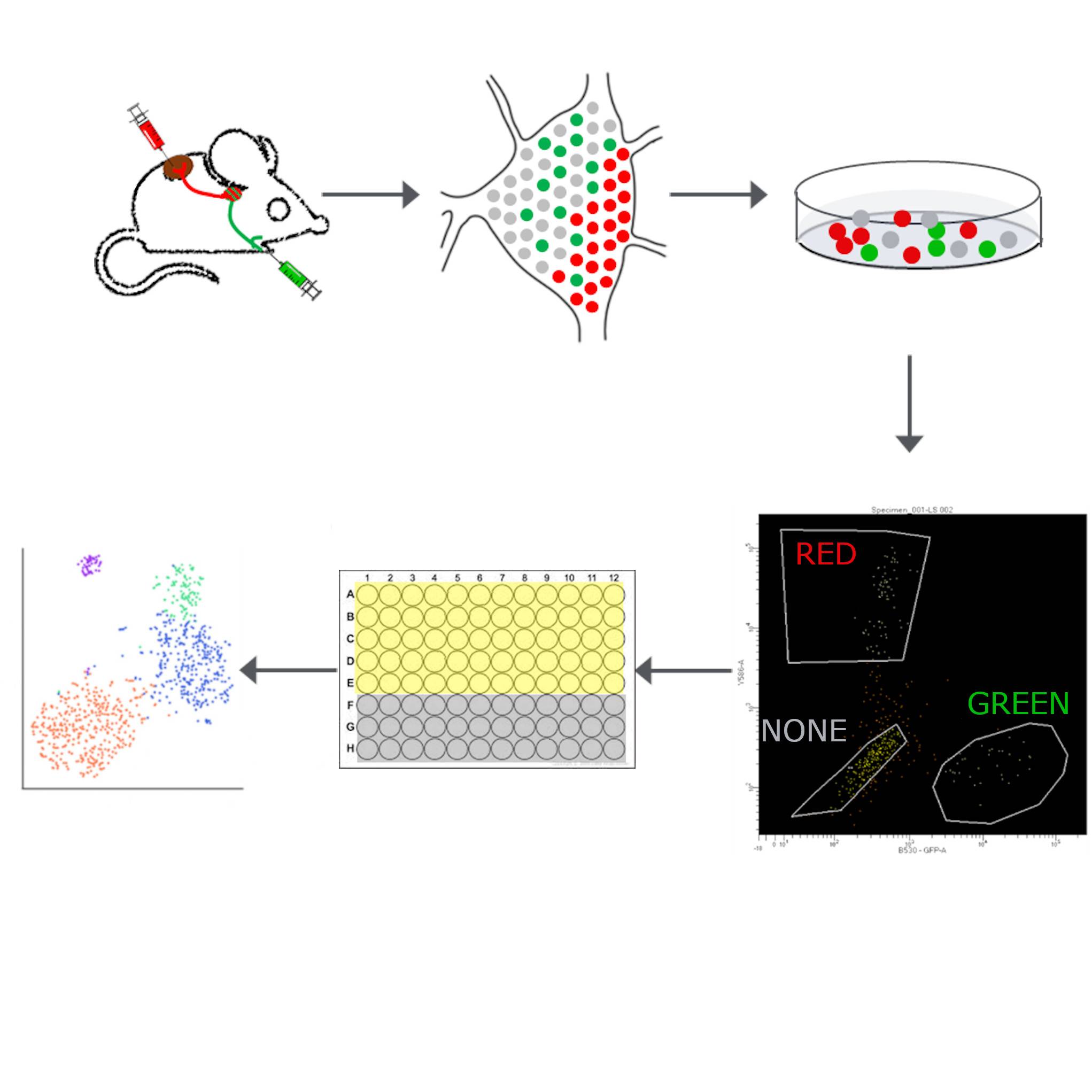
Dataset Overview
Study Purpose: The first scRNA-Seq studies in the peripheral nervous system identified molecular diversity in sympathetic and sensory ganglia. This raised the possibility that target innervation provides a source of molecular diversity in these systems. Here we perform scRNA-Seq in conjunction with retrograde tracing into a sympathetic ganglion. We compared the transcriptional profiles of neurons in the stellate ganglion that project to brown adipose tissue (BAT) vs. forelimb. These targets were chosen because their projections share a similar anatomical trajectory via the intercostal nerves, and they are both involved in thermoregulation. Moreover, sympathetic nervous system (SNS) tone causes vasodilation in BAT, the opposite of what is usually observed.
Data Collection: Cells were sorted based on their relative dye fluorescent and collected in a total of 11 96-well plates and processed in 3 different sets. Pooled, 3’-end cDNA libraries were sequenced on an Illumina NextSeq 500/550 platform. Raw sequencing files (fastq) of each plate are available in the source folder. The reads were aligned to mouse genome, GRCm38, gencode version M13 (Ensembl 88) using STAR v2.5.3a. The aligned reads were assigned to genes using featureCounts v1.5.3. The logs and results from each plate are available in the docs folder. Finally, the bam files, one per plate and available in the primary folder, were sorted and indexed using Samtools v1.4.1 (http://www.htslib.org/), duplicated reads were removed and a UMI count matrix was generated using umi tools. The logs and command input are available in the docs folder, and the final count matrices for each plate are available in the derivative folder. A total of 1039 cells were analyzed.
Primary Conclusion: In contrast to what was seen in sensory neurons, we found no evidence of target-specific transcriptional profiles in sympathetic ganglia in the adult mouse. However, the proportion of the four main neuronal classes varies across targets.
Curator's Notes
Experimental Design: A freshly harvested stellate ganglia were dissociated into single neurons and sorted based on relative fluorescence into 96-well plates. FACSAria sorter with a square cuvette with a 130-μm nozzle at 12 PSI was used. Cells were sorted based on the signal from Alexa 488, Alexa 555, or the lack of any signal. A plate-based single-cell sequencing was performed as described in Snyder et al., 2019 (https://doi.org/10.1126/sciimmunol.aav5581)
Completeness: This dataset is a part of a larger study, "Expression of molecular markers for SG neuronal subpopulations in 3 sympathetic ganglia."
Subjects & Samples: Male (n=6) adult mice (RRID:IMSR_JAX:000664) were used in tis study.
Primary vs derivative data: Primary data is organized into three sets of cDNA libraries and then into the folder with ID of 96 well plates. Each subfolder contains the bam files while raw sequencing files (fastq) of each plate are available in the source folder. The final count matrices for each plate are available in the derivative folder.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Lee, S., & Zeltser, L. (2018). Retrograde labeling of brown adipose tissue (BAT)-projecting sympathetic neurons with cholera toxin B (CTB) v1. https://doi.org/10.17504/protocols.io.wjrfcm6
Lee, S. (2022). Ganglia dissociation and single-cell sorting v1. https://doi.org/10.17504/protocols.io.14egn79e6v5d/v1
Neri, D. (2022). Single-cell sequencing and analysis v1. https://doi.org/10.17504/protocols.io.bp2l61drdvqe/v1
Lee, S., Neri, D., & Zeltser, L. (2022). Single cell RNA sequencing of retrogradely labeled mouse stellate ganglion neuron v1. https://doi.org/10.17504/protocols.io.261generdg47/v1
