Single nucleus sequencing of human C2 dorsal root ganglia tissue from fusion surgeries
Single nucleus RNA sequencing (10x) was performed on 12 dorsal root ganglia from 12 patients undergoing C1-C2 fusion surgery
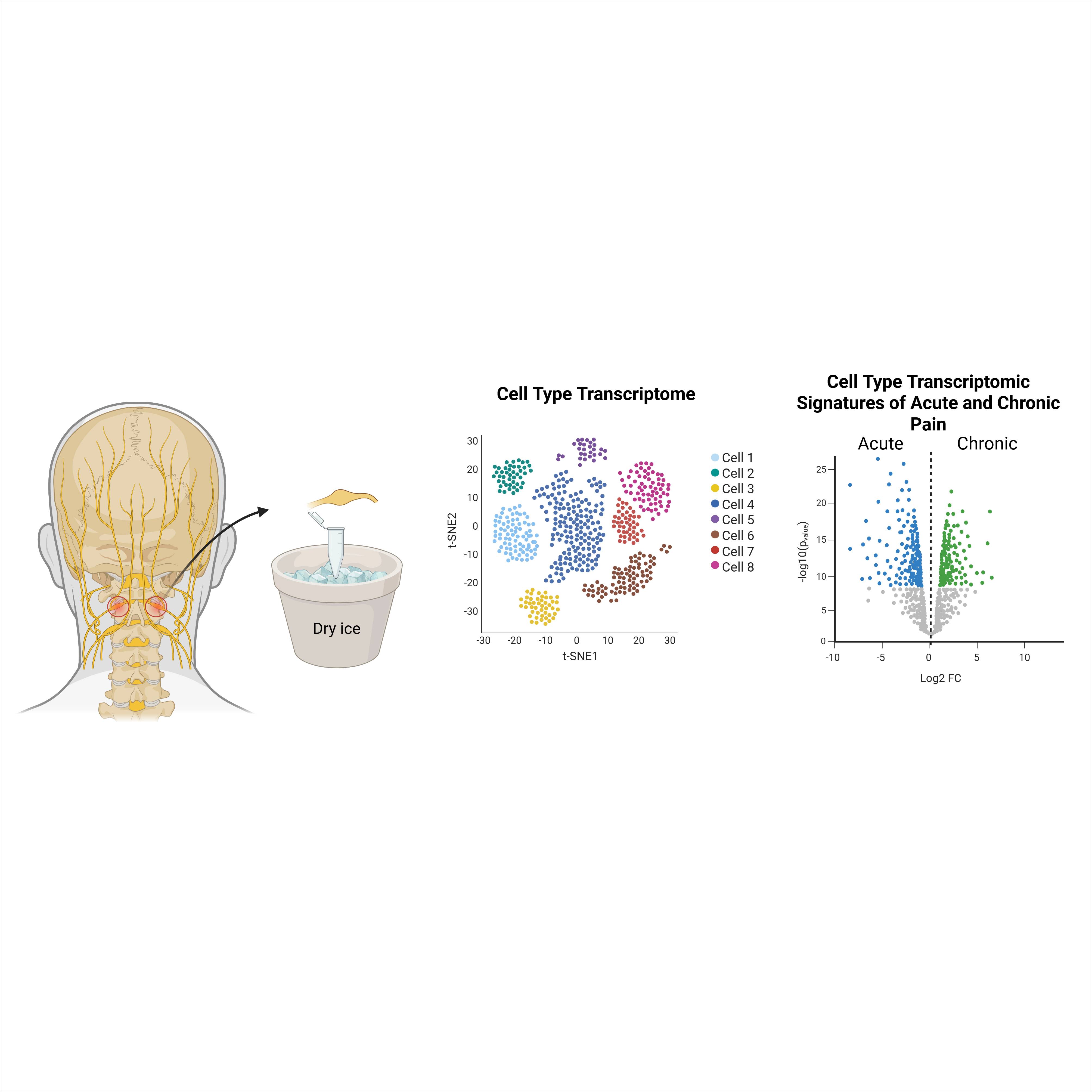
Dataset Overview
Study Purpose: In this study we used single-nucleus RNAseq to characterize the transcriptome of cells in the C2 DRGs and characterize the differences in patients with chronic pain.
Data Collection: 10X Flex single-nucleus RNAseq was performed on 12 C2 DRG collected from 12 patients with acute or chronic pain during atlanto-axial fusion/arthrodesis surgery.
Primary Conclusion: We present a map of the C2 hDRG transcriptome and identified distinct differences associated with chronic pain in non-neuronal cells (endothelial, fibroblast and myelinating Schwann cells).
Curator's Notes
Experimental Design: C2 dorsal root ganglia were collected from patients undergoing atlanto-axial fusion/arthrodesis surgery and immediately flash frozen in the operating room before storage at -80°C. Tissues were shipped to UTD on dry ice for processing. Single cells were harvested and isolated from human DRG tissue, followed by fixation and probe hybridization for 16 hours using the 10X Genomics Chromium Fixed RNA profiling kit. Library preparation was completed according to manufacturer's protocol and sequenced using NextSeq2000. Sequencing data were processed and mapped to the human genome (GRCh38) using 10X Genomics Fix RNA profiling CellRanger multi pipeline version 7.
Completeness: This dataset is a part of a larger study: "Transcriptome of the human C2 dorsal root ganglia in C1-2 arthrodesis surgery: insight for neck pain"
Subjects & Samples: Female (n=2), male (n=10) human patients (ages 53-88 years) with acute (n=7) or chronic (n=5) pain undergoing C1-2 arthrodesis surgery were used in this study.
Primary vs derivative data: Primary data is organized hierarchically by subject folders containing sample folders (sam-UTD-CCXXXX-DRGL/DRGR) with "count" subfolders containing 10X Genomics CellRanger pipeline outputs. Each count folder includes raw and filtered feature-barcode matrices (HDF5 and MTX formats), probe sets, and analysis results including PCA, t-SNE, UMAP projections, clustering results, and differential expression analysis. There is no derivative data folder.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Sankaranarayanan, I., M Mwirigi, J., B Lesnak, J., Saad Yousuf, M., & Price, T. (2024). Single-cell RNA sequencing from human dorsal root ganglion v2. https://doi.org/10.17504/protocols.io.4r3l2qpr4l1y/v1
P Hofstetter, C., Tran, N., & Payne, C. (2024). Human Tissue Procurement from Atlanto-Axial Fusion Surgery v1. https://doi.org/10.17504/protocols.io.eq2lywoervx9/v1
Curatolo, M., Payne, C., Chiu, A. P., Tran, N. T., Yap, N., Hofstetter, C. P., Lesnak, J. B., Arendt-Tranholm, A., Price, T. J., Jarvik, J. G., & Turner, J. A. (2025). Patient phenotyping for molecular profiling of neck and low back pain – Study protocol. Neurobiology of Pain, 18, 100186. https://doi.org/10.1016/j.ynpai.2025.100186
Described by
Arendt-Tranholm, A., Sankaranarayanan, I., Payne, C., Mancilla Moreno, M., Mazhar, K., Yap, N., Chiu, A. P., Barry, A., Patel, P. J., Inturi, N. N., Tavares-Ferreira, D., Amin, A., Karandikar, M., Jarvik, J. G., Turner, J. A., Hofstetter, C. P., Curatolo, M., & Price, T. J. (2025). Transcriptome of the human C2 dorsal root ganglia in C1-2 arthrodesis surgery: insight for neck pain. Brain. https://doi.org/10.1093/brain/awaf370
