Quantitation of nodose neurons labelled by retrograde tracing from selective gastric mucosal and gastric muscle injection
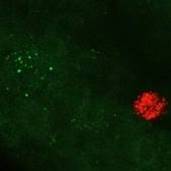
Microinjection of fluorescent nanospheres into either the muscle or mucosal of the stomach in the corpus regions in rats labels distinct populations of nodose neurons
Dataset Overview
Study Purpose: In this study, we have developed methods to identify individual vagal sensory neurons that project to and innervate specific tissues within the stomach and send information to the brain so that their properties and roles in gastric function can be studied in detail.
Data Collected: Confocal z-stacks (three-dimensional images) of the whole nodose ganglion showing the positions and sizes of labeled neurons were generated. From these images, labeled neurons were identified and counted, and their sizes quantified so that the properties of different populations could be compared.
Primary Conclusion: Nodose neurons innervating specific tissues in the stomach can be reliably identified using our methods. Relatively small populations of nodose neurons project to any one section of the stomach wall, but separate populations innervate the muscle and mucosa in these regions. The population of nodose neurons projecting to the gastric mucosa includes many large neurons, while muscle afferent neurons include many small neurons.
Curator's Notes
Experimental Design: Neurons in the nodose ganglion were labelled using retrograde tracers injected into either the mucosal layers or the muscle layers or both in the corpus region of the rat stomach. The injection sites in stomach samples are dissected out and mounted on microscope slides in wells constructed using rubber take to create wells for imaging thick tissue and cover-slipped. Injection sites were then scanned at low power using a confocal fluorescence microscope (Zeiss LSM800 Airyscan). Large tile scans were collected at various focal planes (z-stack) in order to cover the entire ganglia in both red and green channels. Identification of tracer labeled nodose neurons as well as determination of their cell body area size are determined using Zen Blue 3.0 software (Zeiss).
Completeness: This dataset is part of a larger study: "Mapping Stomach Autonomic Circuitry and Function"
Subjects & Samples: Female (n=6) and male (n=9) adult Sprague-Dawley rats (RRID:RGD_8553001) were used in this study.
Primary vs derivative data: The primary data folder is structured with subfolders organized by subject ID and then sample ID, respectively. Each sample subfolder contains microscopy imaging files in the .czi format. In the derivative data folder, an Excel file provides a summary of all data points measured from each sample. Image data in JPEG2000 and OME-TIFF formats was derived from primary images in the .czi format. MBF Bioscience converted primary images to JPEG2000 (.jpx) with a 20:1 compression ratio for web streaming and visualization on the SPARC Data Portal. Furthermore, primary images were also converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Furness, J., & Stebbing, M. (2019). Protocols for selective injection of tracer into the gastric mucosa and the gastric muscle of rat v1. https://doi.org/10.17504/protocols.io.yxmvmxko9l3p/v1
