RNAseq analysis of nodose neurons projecting to specific regions and tissue layers within the rat stomach
Vagal sensory neurons projecting to either the muscle or mucosal layers of the rat stomach in the fundus, corpus or antrum were identified using localized microinjections retrograde tracer, before being isolated and characterized using RNAseq.
Dataset Overview
Study Purpose: To sequence in depth and compare the transcriptomes of vagal afferent neurons projecting to different tissues and different regions of the rat stomach.
Data Collected: The data consists of:
- raw RNAseq data analysis and quality control analysis for each sample present in the 'primary/[subject]/[sample]/RNAseq' folders
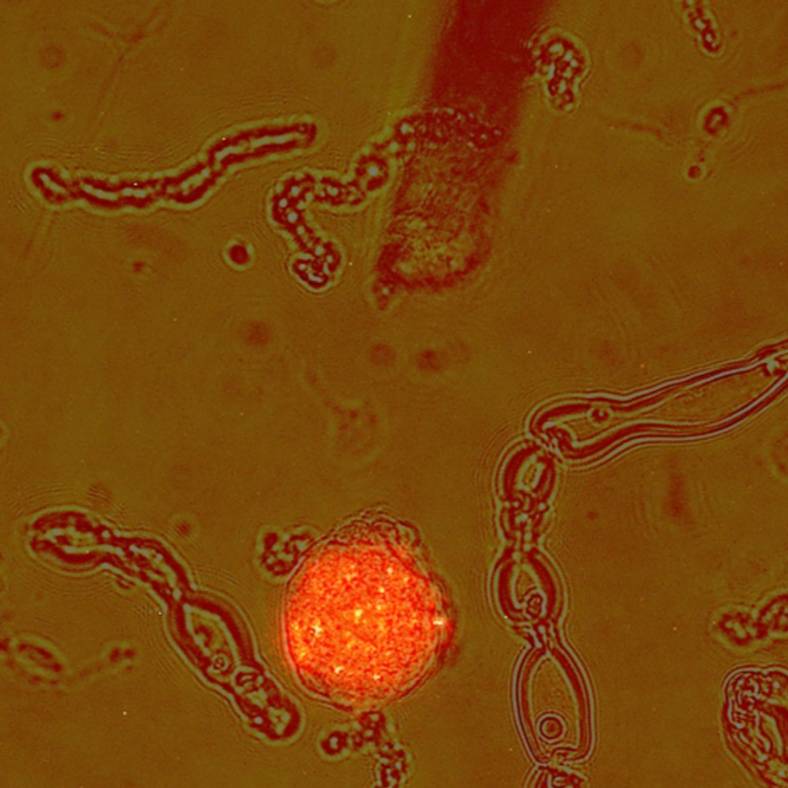
- images of the dissociated cells (brightfield and fluorescence) showing labeled and unlabelled cells in the 'primary/[subject]/[sample]/Microscopy' folders
- low magnification images of the tracer bead injections sites in the stomach in the 'primary/[subject]/[sample]-stomach/Macroscopy' folders
- micrographs of stomach sections through injection sites used to determine the extent of bead infiltration within the stomach tissues in the 'primary/[subject]/[sample]-stomach/microscopy' folders
Primary Conclusion: QC analysis of samples and raw results files confirmed that high quality mRNA and sequencing data was obtained. Targeted analyses of genes known to be functionally important in gastric vagal afferent neurons confirmed the specificity of our tracing methods and revealed clear differences in gene expression based on the gastric region and gastric tissue innervated. The dataset represents an important resource for further analysis and discovery. We expect it to facilitate a deeper, more detailed understanding of the role of gastric afferent neurons in the functionally diverse gastric regions studied.
Experimental Design: Neurons in the nodose ganglion were labeled using retrograde tracers injected into either the mucosal layers or the muscle layers or both in the corpus, antrum, and fundus regions of the rat stomach. After allowing time for the tracer to be transported back to vagal afferent cell bodies, the nodose ganglia were harvested and dissociated from at least 6 animals (3 male and 3 female) per region and tissue. Labeled neurons were identified using a fluorescence microscope and harvested individually via a micropipette. Pooled samples of small numbers of neurons were sequenced using a short read (75bp), single-ended method with at least 25 million reads per sample so that expression of low copy number genes could be detected.
Completeness: This dataset is part of a larger study: "High resolution retrograde tracing combined with detailed molecular analyses to characterize the subpopulations of vagal afferent neurons monitoring the functional state of the stomach in rats"
Subjects & Samples: Male (n=16) and female (n=16) Sprague-Dawley rats (RRID:RGD_10395233) between 9 and 22 weeks old were used in the study.
Primary vs derivative data: Primary folder contains raw RNA sequencing data (.fastq.gz), microscopy (.czi), and macroscopy images (.jpg) in subfolders nested under folders organized by the subject ID. Derivative folder contains an excel spreadsheet with aggregated data on histological analysis of injection sites and an automated data viewer in the form of an excel spreadsheet that allows the sequencing data to be interrogated in relation to any rat gene. Quality control data from automated electrophoretic analysis of samples after mRNA amplification and before sequencing is in the source folder.
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Stebbing, M. J., Molero, J. C., & Furness, J. B. (2022). High resolution retrograde tracing, morphological analysis, RNA sequencing and differential expression analysis of identified vagal afferent neurons innervating the muscle or mucosa in 3 different stomach regions in the rat. The FASEB Journal, 36(S1). Portico. https://doi.org/10.1096/fasebj.2022.36.s1.r4114
Is Supplemented by
Furness, J., & Stebbing, M. (2019). Protocols for selective injection of tracer into the gastric mucosa and the gastric muscle of rat v1. https://doi.org/10.17504/protocols.io.yxmvmxko9l3p/v1
