Mapping of intrinsic cardiac nervous system (ICN) neurons in a 3D reconstructed rat heart
Single intrinsic cardiac nervous system (ICN) neurons were marked in an image volume of heart sections that was anatomically delineated for the 3D reconstruction of the whole heart with mapped locations of individual ICN neurons.
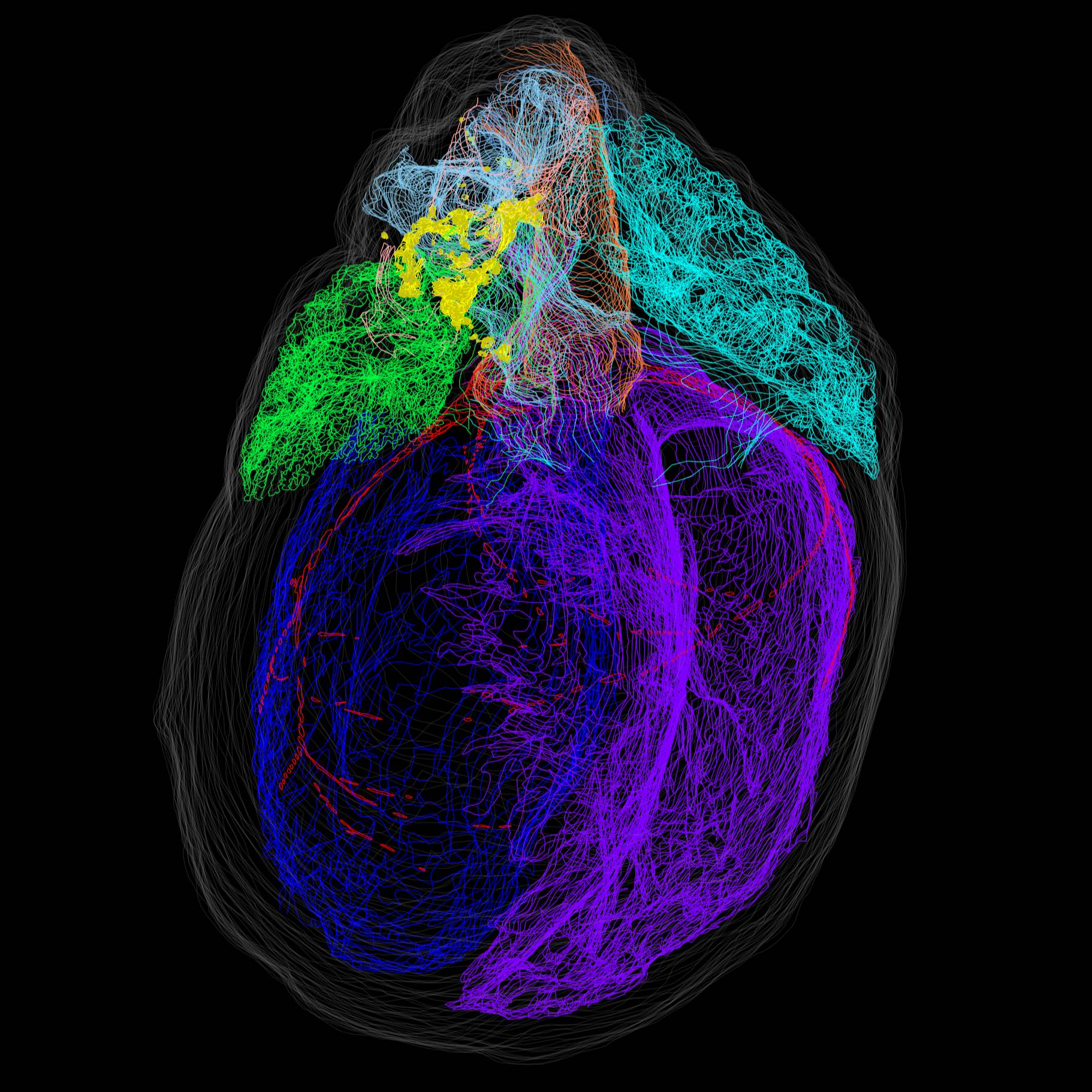
Dataset Overview
Study Purpose: The purpose of this study was to establish a comprehensive 3D atlas of the rat intrinsic cardiac nervous system (ICN) at the single-cell resolution to characterize the distribution patterns of intrinsic cardiac neurons between individuals and across sexes. To this end, we used a combination of high-throughput sectioning and imaging with a Knife Edge Scanning Microscope (KESM) to assemble image volumes. Tissue Mapper software (MBF Bioscience) to map ICN neurons and to annotate key cardiac anatomy for 3D heart reconstructions.
Data Collection: The heart was fixed and then whole-mount diffusion stained with Cresyl Echt Violet to visualize ICN neurons. The heart was then sectioned and imaged using a Knife Edge Scanning Microscope (KESM) to produce image stacks. Single neurons were mapped, and cardiac anatomy was segmented on select sections of the image stacks with the Tissue Mapper software. The spatial distribution of individually marked neurons and annotated heart anatomy were then visualized using a 3D reconstructed model of the heart.
Primary Conclusion: None stated
Curator's Notes
Experimental Design: The rat heart was first fixed and stained with Cresyl Echt Violet to visualize the ICN neurons. The whole hearts were then sectioned and imaged in three different orientations with a Knife Edge Scanning Microscope (KESM). The resulting image tiles were stitched together to assemble 7 image stacks. Afterward, single neurons were mapped, and cardiac anatomy was segmented on select sections of the image stacks with Tissue Mapper software (from MBF Bioscience). The spatial distribution of individually marked neurons and annotated heart anatomy were then visualized in a 3D reconstructed model of the heart.
Completeness: Batch
Subjects & Samples: This study used one normal male Fischer 344 rat.
Primary vs derivative data: The primary folder contains microscopic images (jpx). The derivative folder contains segmentation data (xml) of mapped ICN neurons and cardiac anatomy delineations. Segmentation data was created in and can be viewed in MBF Bioscience software.
Important Notes: This dataset was a preliminary research study for a larger study of additional rat hearts (https://doi.org/10.26275/yh5c-5pjy).
Code Availability: Not Applicable
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Leung, C., Heal, M., Robbins, S., Moss, A., Monteith, C., & Tappan, S. (2020). Single-Cell ICN Neuron Mapping and 3D Heart Reconstruction with Tissue Mapper v1. https://doi.org/10.17504/protocols.io.bdz5i786
Described by
Achanta, S., Gorky, J., Leung, C., Moss, A., Robbins, S., Eisenman, L., Chen, J., Tappan, S., Heal, M., Farahani, N., Huffman, T., England, S., Cheng, Z. (Jack), Vadigepalli, R., & Schwaber, J. S. (2020). A Comprehensive Integrated Anatomical and Molecular Atlas of Rat Intrinsic Cardiac Nervous System. IScience, 23(6), 101140. https://doi.org/10.1016/j.isci.2020.101140
