Quantitation of nodose neurons labelled by retrograde tracing from selective injections into the gastric mucosa or muscle in the antrum and fundus
Microinjections of a non-diffusible retrograde tracer were made into the muscle layer or the mucosal layer of the rat stomach in either the antrum or the fundus region. Retrogradely labelled nodose neurons were studied using 3-D confocal microscopy.
Dataset Overview
Study Purpose: In a previous study we have developed methods to identify individual vagal sensory neurons that project to and innervate specific tissues within the stomach and used those methods to look at the vagal afferent innervation of the corpus region. In this study we apply these methods to identify and study the vagal afferent neurons that innervate the mucosa or the muscle layers in the antrum and fundus regions of the rat stomach, which are functionally and structurally distinct from the corpus region.
Data Collected: Neurons in the nodose ganglion were labelled using fluorescent retrograde tracer beads injected into either the mucosal layers or the muscle layers in the antrum region or the fundus region of the rat stomach. The tracer injection sites were examined in cryostat sections of the stomach to confirm correct targeting of the tracer beads. Confocal z-stacks (three dimensional images) of the whole nodose ganglion showing the positions and sizes of labelled neurons were generated. From these images, labelled neurons were identified, counted and their sizes quantified so that properties of different populations could be compared.
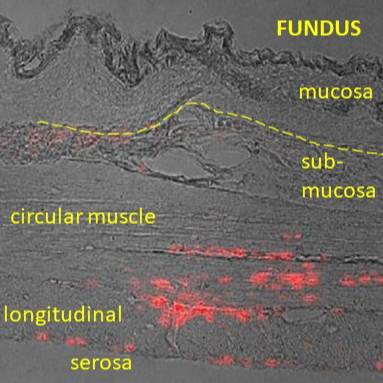
Primary Conclusion: Specific tracer injections into the antrum mucosa and antrum muscle were accurately and consistently targeted, based on histological analysis of the stomach injection sites. Beads injections into the fundus muscle did not enter mucosal layers, but injections into the thin fundus mucosa were not confined to the mucosa, so that specific targeting of the neurons innervating this tissue layer in the fundus was not possible. As for the corpus region, the nodose neurons projecting to the antral gastric mucosa were included many large neurons (based on cross sectional area), while muscle afferent neurons include many small neurons, suggesting these populations of neurons are distinct. Neurons labelled from injections into the fundus muscle were also in the small size range, but neurons labelled from fundus mucosa injections were, on average, intermediate in cross sectional area, supporting the view that both mucosa innervating and muscle innervating neurons were labelled by the mucosa injections.
Curator's Notes
Experimental Design: This dataset contains quantitative confocal microscopy data rat nodose neurons labeled by specific retrograde tracer injections within the stomach, i.e. either the muscle layers or mucosal layer in the fundus or antrum. These neurons were identified using retrograde tracing injections (red retrobeads) in rats 1-2 weeks before nodose ganglia were harvested after perfusion of the animal with fixative, carefully dissected squash mounted on slides and imaged in whole ganglia using confocal microscopy.
Completeness: This dataset is a part of a larger study: "Mapping Stomach Autonomic Circuitry and Function"
Subjects & Samples: Male (n-8) and female (n=1) rats (RRID:RGD_10395233) were used in this study.
Primary vs derivative data: Primary data is organized in folders by subject ID and sample ID respectively. Each sample folder consists of:confocal tile scan z-stacks of the whole nodose ganglia showing labeled cells;low magnification images of the tracer bead injection sites in the stomach in the 'primary/[subject]/[sample]-stomach/Macroscopy' folders, and micrographs of stomach sections through injection sites used to determine the extent of bead infiltration within the stomach tissues in the 'primary/[subject]/[sample]-stomach/microscopy' folders. The derivative data folder consists of an Excel spreadsheet with aggregated, qualitative data on histological analysis of injection sites, quantitative data on the numbers of cells labeled and the sizes of those cells determined from confocal z stacks by manually tracing the perimeter of labeled cells at image plane where their cross-sectional area was largest, using zen software. Image data (JPEG2000 and OME-TIFF) was derived from primary images (.tif). Primary images were converted with lossless compression to JPEG2000 (.jpx) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal. Primary images were also converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplement to
Molero, J., Stebbing, M., & Furness, J. (2024). Quantitation of nodose neurons labelled by retrograde tracing from selective gastric mucosal and gastric muscle injection [Data set]. In Mapping Stomach Autonomic Circuitry and Function (Version 1). SPARC Portal. https://doi.org/10.26275/GUD3-ALHY
