Morphologies dimensions and targets of gastric nitric oxide synthase neurons in the rat stomach
This dataset contains images and analysis used to quantify the distributions and targets of neuronal nitric oxide synthase (nNOS) immunoreactive neurons in the rat stomach.
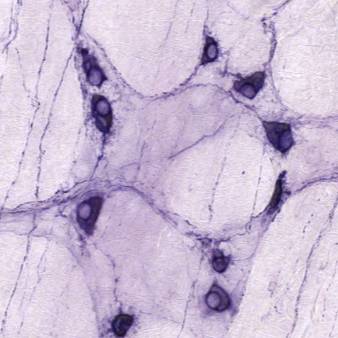
Dataset Overview
Study Purpose: To investigate the distributions and targets of neuronal nitric oxide synthase (nNOS) neurons in the muscle layers of the rat stomach in order to better understand how these neurons influence stomach movements.
Data Collected: This dataset contains images and analysis used to quantify the distributions and targets of neuronal nitric oxide synthase (nNOS) immunoreactive neurons in the rat stomach.
Primary Conclusion: These data show that nitrergic neurons make up about 30% of all myenteric neurons in all gastric regions, with a small population appearing in the submucosal ganglia. The myenteric cell bodies had single axons, Type I morphology, and a wide range of sizes. Five targets were identified: the longitudinal, circular, and oblique layers of the external muscle, the muscularis mucosae, and arteries within the gastric wall. There was also a dense innervation of the pyloric sphincter. Both nNOS immunohistochemistry and NADPHd histochemistry showed that nitrergic neurons did not provide baskets of terminals around myenteric neurons, which suggests gastric interneurons do not express nNOS, unlike intestinal interneurons. The high densities of nNOS fibers throughout the CM and the pyloric sphincter suggest there is highly regulated, fine graded inhibitory control of motor function in these regions.
Curator's Notes
Experimental Design: Stomachs from adult Sprague-Dawley rats were collected, all three main regions of the stomach (fundus, corpus, and antrum) along the lesser and greater curvature were investigated. Wholemounts and sections (12μm) were used to investigate the distributions of nNOS neurons and their targets. Immunohistochemical staining with anti-nNOS, anti-Hu, and anti- α smooth muscle actin antibodies and NADPH diaphorase (NADPHd) histochemistry staining were utilized. Neurons were imaged using fluorescent microscopy, confocal microscopy, and a slide scanner to document the staining in large regions of the stomach wall. Neurons were drawn manually using camera lucida to document differences in morphology.
Completeness: This dataset is part of a larger study: "Mapping Stomach Autonomic Circuitry and Function"
Subjects & Samples: Female (n=16) and male (n=9) adult Sprague-Dawley rats (RRID:RGD_8553001) were used in this study.
Primary vs derivative data: The primary data folder is organized into subfolders by subject ID and then sample ID, respectively. Within each sample subfolder, you will find microscopy imaging files in the .czi format, accompanied by a text file describing the data files present in that directory. Additionally, digital camera images of the rat stomach are included in a compressed (.zip) format. The derivative data folder comprises quantitative data obtained from tissue stained with nNOS (neuronal nitric oxide synthase) labeled with AlexaFluor 488 and Hu C/D (pan-neuronal marker) labeled with AlexaFluor 594. Image data, available in both JPEG2000 and OME-TIFF formats, was derived from the primary images stored in the .lif format. MBF Bioscience performed the conversion of primary images to JPEG2000 (.jpx) using a 40:1 compression ratio for web streaming and visualization on the SPARC Data Portal. Additionally, the primary images were converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience. The ses-NADPHd-xxx.pdf files in the source folder contain the raw scanned images of hand-drawn neurons before separation and labeling as individual cells in the (ses-NADPHd)>(drawing).
Files
0 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Di Natale, M. R., Hunne, B., Liew, J. J. M., Fothergill, L. J., Stebbing, M. J., & Furness, J. B. (2022). Morphologies, dimensions and targets of gastric nitric oxide synthase neurons. Cell and Tissue Research, 388(1), 19–32. https://doi.org/10.1007/s00441-022-03594-0
Is Supplemented by
Di Natale, M., JL Liew, J., Hunne, B., Stebbing, M., & Furness, J. (2022). Immunohistochemistry in wholemounts and cryostat sections in rat stomach v1. https://doi.org/10.17504/protocols.io.yxmvmnk5og3p/v1
Di Natale, M., Hunne, B., Stebbing, M., & Furness, J. (2022). Quantification of rat stomach surface area v1. https://doi.org/10.17504/protocols.io.6qpvr6dmbvmk/v1
Di Natale, M., Hunne, B., JL Liew, J., Fothergill, L., Stebbing, M., & Furness, J. (2022). NADPH diaphorase histochemistry in rat gastric nitric oxide synthase neurons v1. https://doi.org/10.17504/protocols.io.eq2lyn3eevx9/v1
Hunne, B., Furness, J., Stebbing, M., & Fothergill, L. (2021). Clearing-enhanced 3D (Ce3D) clearing method, immunohistochemistry and quantitation of rat gastric enteric ganglia v1. https://doi.org/10.17504/protocols.io.bp2l6b845gqe/v1
