Michigan High Res EEG - UMHS 0023
William C Stacey, M.D.
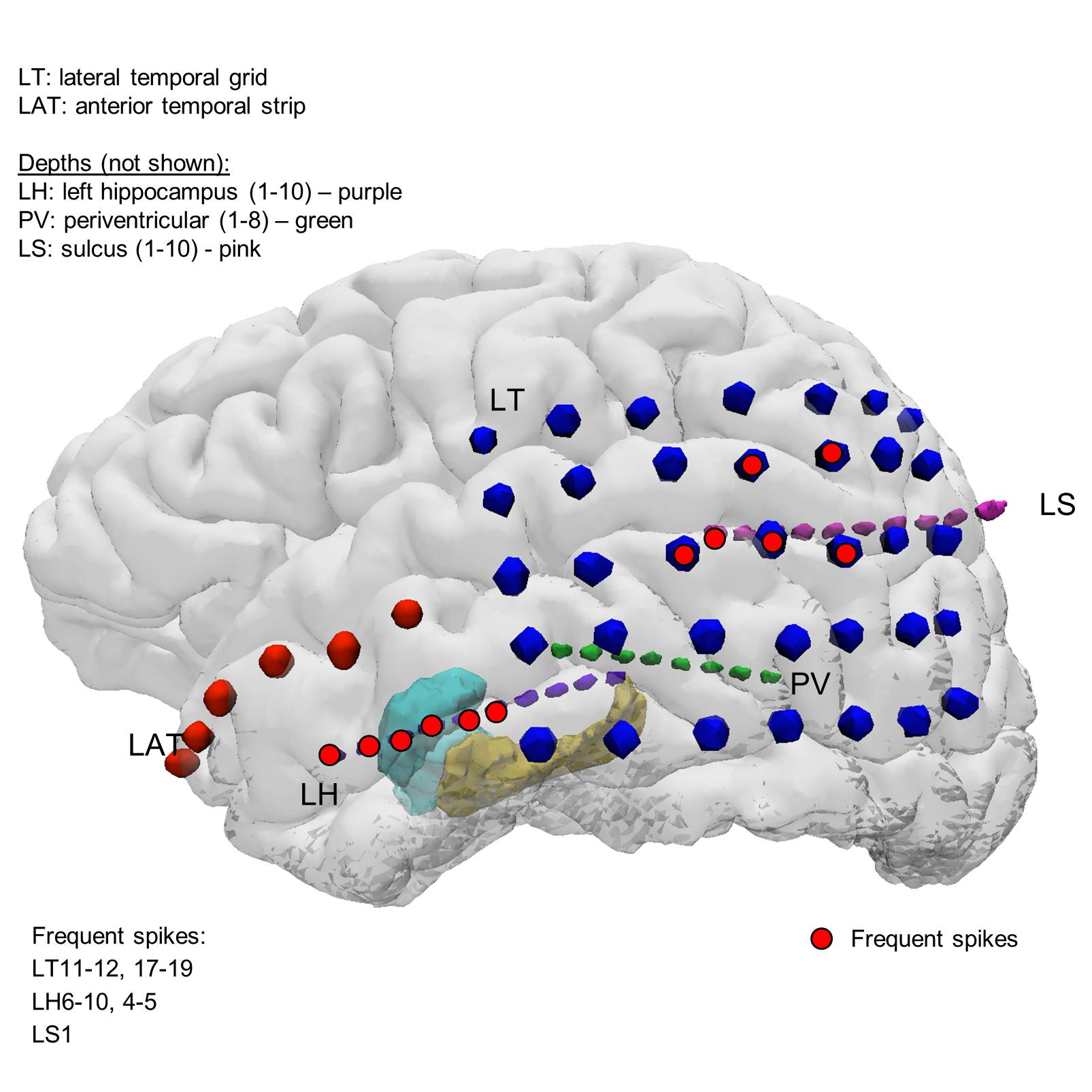
Scalp, intracranial grid, and standard depth EEG for UMHS0023. Neuropace placed.
Dataset Overview
Overview
29 y/o man with 8 days of scalp EEG, standard depth EEG (28 electrodes), intracranial grid EEG (41 electrodes), and sleep staging. Patient had Neuropace placed instead of resection.
The electrodes in the seizure onset zones are: [ 60:62 45:51 5:7 10:13 15:19 23:26 29:31 ].
File Information
EEG data and metadata are stored in the files.
TXT files
- database-patientID-sessionNumber.chanHdr.txt: Meta data on the channels used
- nChan: Number of active channels
- nChanTot: Total number of channels
- macro: Macroelectrodes
- micro: Microelectrodes
- depth: Intracranial grid electrodes
- ECoG: Intracranial grid electrodes
- external: Scalp electrodes
- chanMap: Electrodes indicated in the channel map
- database-patientID-sessionNumber.clinHdr.txt: Clinical meta data
- knowSOZ: Whether the seizure onset zone was known
- knowIfResected: Whether there will be resection
- didResect: Whether there was resection
- knowResectedVol: Whether the resected volume was known
- knowOutcome: Whether the patient oucome was known
- ILAE_outcome: Epilepsy surgery seizure outcome using the International League Against Epilepsy (ILAE) Outcome Scale
- RV: Electrodes placed in the resected volume
- SOZ: Electrodes placed in the seizure onset zone
- highIIS: Electrodes placed at locations with interictal spikes
DOCX files
- database-patientID.LTM.docx: De-identified copy of clinical and hospitalization record
- database-patientID.surgical-report.docx: Surgical report
- database-patientID.pathology.docx: Pathology information
- database-patientID.electrode-placement.docx: Electrode placement
PPT files
These files contain clinical information.
LAY files
These files contain Persyst EEG data.
DAT files
These files contain Persyst EEG data.
MATLAB code
- MAT files contain EEG metadata.
MinutesFromStart.matcontains the duration (minutes) from the first EEG sample of the patient to the first EEG sample taken in each session.PatientID-sleep-scores.matcontains information from sleep staging.- time: Time at the sleep stage
- stageCode: The indices that correspond to the sleep stages in stage
- stage: Sleep stage
- nonREMsleep: Details during non-REM sleep
- LAY files can be read with
inifile.m,lay_data_read.m, andlay_hdr_read.m.
Usage
- Ensure that the codes are in the active folder or in your MATLAB path.
- Change the false to true if you want to read the annotations into the header. Not recommended for long clinical EEG.
- The startIdx and stopIdx are the indexes from the .dat file you are interested in. For the start of the file, startIdx=1. For the end of the file, stopIdx = hdr.datapoints.
hdr = lay_hdr_read(filename,false);
data = lay_data_read(hdr,startIdx,stopIdx);
Files
Root Directory
1 - 0 of 0 files
About this dataset
Publishing history
December 18, 2023
Originally Published
December 18, 2023 (Version 1)
Last Updated
Cite this dataset
Stacey, W. C. (2023). Michigan High Res EEG - UMHS 0023 (Version 1) [Dataset]. Pennsieve Discover. https://doi.org/10.26275/S3JN-EO59
Formatted as: | | | More on Crosscite.org
