Validated computational models predict vagus nerve stimulation thresholds in preclinical animals and humans
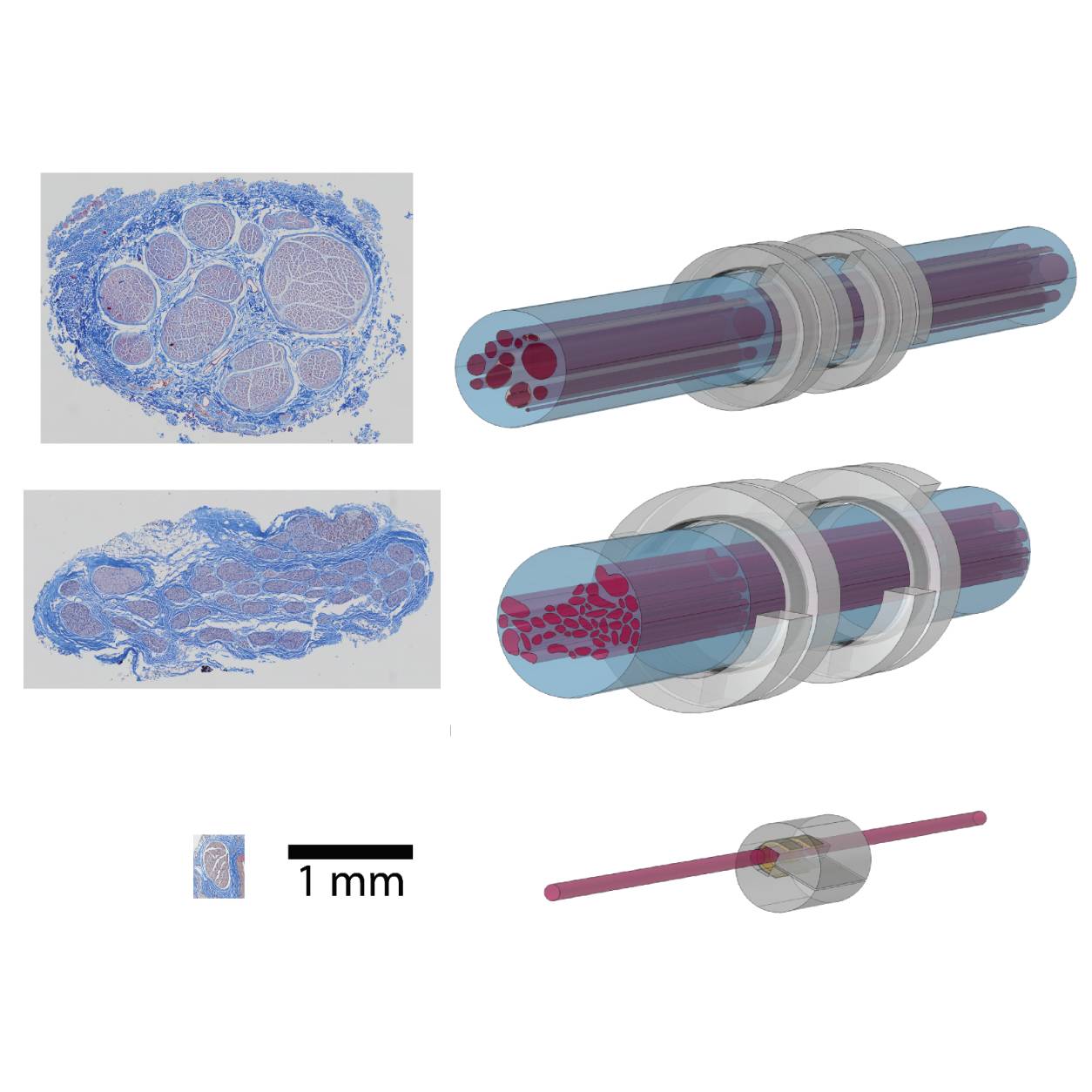
Validated computational models of rat (Micro-Leads Neuro, n=1), pig (LivaNova, n=20), and human (LivaNova, n=9) vagus nerve stimulation, built with ASCENT v1.1.1, using histology to define nerve morphologies, and using the cuff electrodes and waveforms.
Dataset Overview
Study Purpose: To implement, simulate, and validate computational models of VNS across rats, pigs, and humans for a range of fiber types and stimulation pulse widths.
Data Collection: We used ASCENT v1.1.1 to build a 3D model of nine human (acute and chronic; LivaNova), 20 pig (acute; LivaNova), and 1 rat (acute; Micro-Leads Neuro) vagus nerves and cuff electrodes. We simulated thresholds for myelinated and unmyelinated fibers for biphasic rectangular pulses to match stimulation-evoked physiological and neural responses from in vivo and clinical studies.
Primary Conclusion: Modeled activation thresholds quantitatively matched stimulation-evoked responses from preclinical and clinical studies for a large range of nerve morphologies, fiber types, and stimulation pulse widths.
Curator's Notes
Experimental Design: Computational models of vagus nerve stimulation in rats, pigs, and humans.
Completeness: This dataset is complete.
Subjects & Samples: Only virtual subjects were used in this study, which are based on previously published histology from perfused rats, fresh pigs, and embalmed human cadavers.
Primary vs derivative data: Not applicable. This is a computational study. The files in primary/sub-*/sam-*-sub-*/samples/*/ are ASCENT input configuration files (i.e., sample.json, models/*/model.json) and outputs (i.e., individual fiber activation thresholds models/*/sims/*/n_sims/*/data/outputs/thresh_inner*_fiber*.dat).
Additionally, the files in primary/sub-*/sam-*-sub-*/samples/*/ contain key intermediate files created by ASCENT, including the extracellular potentials applied to the fiber models (models/*/sims/*/n_sims/*/data/inputs/inner*_fiber*.dat) and the stimulation waveform (models/*/sims/*/n_sims/*/data/inputs/waveform.dat).
Code Availability: The code used to parameterize, implement, and simulate the computational models is available in ASCENT v1.1.1 (repository; documentation). To analyze and visualize the data, plotting scripts are located in files/code/, as further detailed in the README.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Musselman, E. D., Pelot, N. A., & Grill, W. M. (2023). Validated computational models predict vagus nerve stimulation thresholds in preclinical animals and humans. Journal of Neural Engineering, 20(3), 036032. https://doi.org/10.1088/1741-2552/acda64
