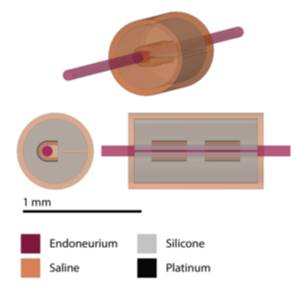
Computational model of vagus nerve stimulation in mice with bipolar cuff electrode
Computational model of vagus nerve stimulation in mice with a bipolar cuff electrode, built with ASCENT v1.1.4, using a generalized nerve morphology.
Dataset Overview
Study Purpose: To implement, simulate, and analyze a computational model of mouse VNS with the MicroLeads bipolar cuff electrode to complement in vivo experiments which recorded laryngeal EMG and heart rate during stimulation.
Data Collection: We used ASCENT v1.1.4 to build a 3D model of a mouse vagus nerves instrumented with the MicroLeads bipolar cuff electrode. We simulated thresholds for 2-5 um “B” and 7-11 um “A alpha” fibers, which we compared to heart rate and laryngeal EMG in vivo responses, respectively.
Primary Conclusion: The model accurately predicted thresholds for A fiber onset/saturation and B fiber onset, and underestimated B fiber saturation. Recruitment order of the fibers was driven by longitudinal fiber alignment for A fibers and fiber diameter for B fibers.
Curator's Notes
Experimental Design: A computation model of vagus nerve stimulation in mice with a bipolar cuff electrode.
Completeness: This dataset is part of a larger study: "Measuring and modeling the effects of vagus nerve stimulation on heart rate and laryngeal muscles"
Subjects & Samples: Only virtual subjects were used in this study.
Primary vs derivative data: Not applicable. This is a computational study. The files in primary/sub-*/sam-*-sub-*/samples/*/ are ASCENT input configuration files (i.e., sample.json, models/*/model.json) and outputs (i.e., individual fiber activation thresholds models/*/sims/*/n_sims/*/data/outputs/thresh_inner*_fiber*.dat).
Additionally, the files in primary/sub-*/sam-*-sub-*/samples/*/ contain key intermediate files created by ASCENT, including the extracellular potentials applied to the fiber models (models/*/sims/*/n_sims/*/data/inputs/inner*_fiber*.dat) and the stimulation waveform (models/*/sims/*/n_sims/*/data/inputs/waveform.dat).
Code Availability: Run the plotting scripts in the dataset using ASCENT v1.1.4 (repository; documentation): see README.txt for details.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Huffman, W. J., Musselman, E. D., Pelot, N. A., & Grill, W. M. (2023). Measuring and modeling the effects of vagus nerve stimulation on heart rate and laryngeal muscles. Bioelectronic Medicine, 9(1). https://doi.org/10.1186/s42234-023-00107-4
