Mapping of the vagal afferent innervation of the mouse lung
Mapping of the vagal afferent innervation of the mouse lung using a combination of viral and genetic approaches to label unique vagal sensory subsets innervating the lung
Dataset Overview
Study Purpose: To study lung innervation of vagal sensory afferent nerves.
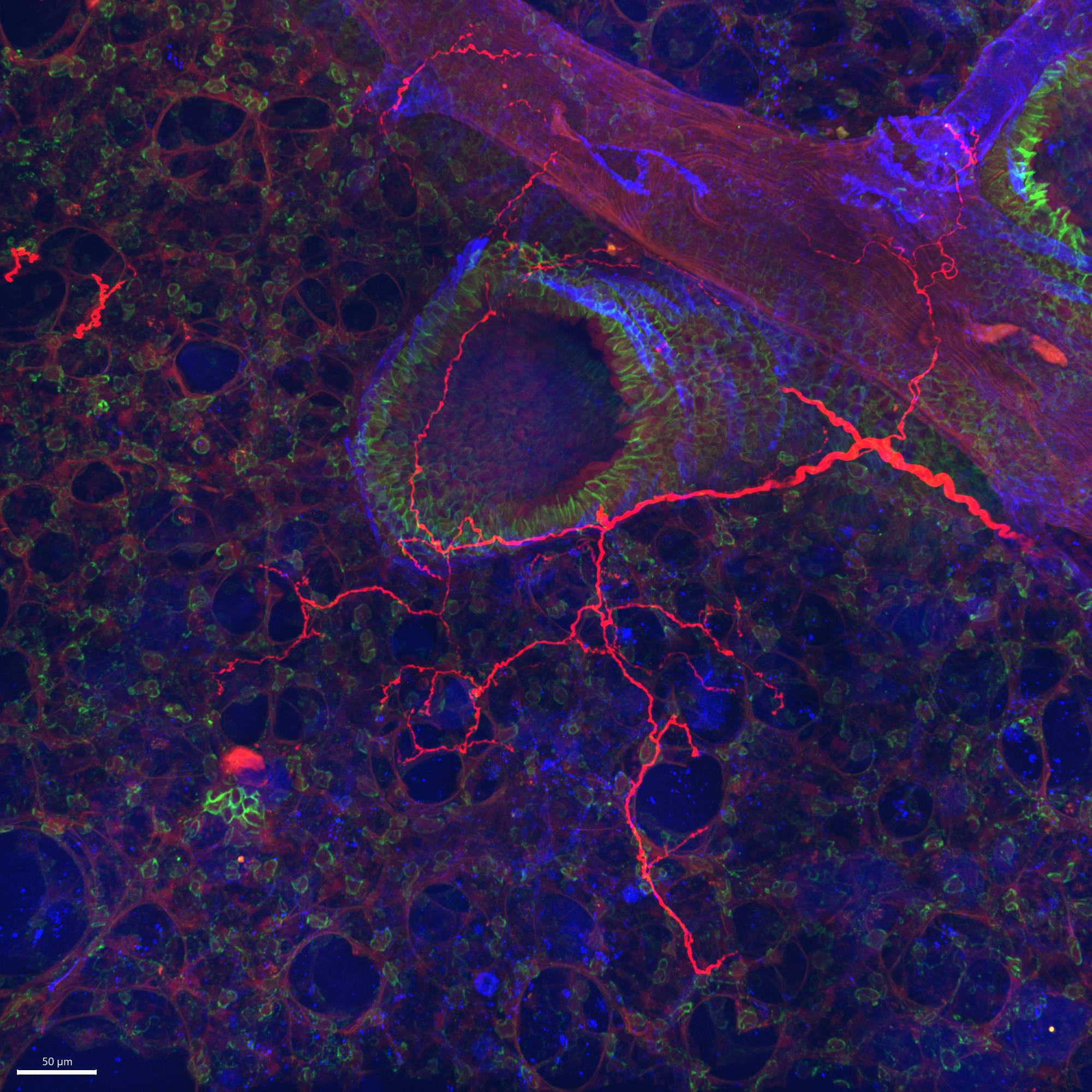
Data Collection: This dataset provides confocal images of lung sections from TRPV1-cre and Tac1-cre mice following injections into the vagal ganglia with an AAV9 vector (flex-GFP) that expresses the fluorescent protein GFP in a cre-dependent manner. Thus the dataset identifies the specific vagal afferents innervating the lung that express TRPV1 (nociceptors) and Tac1 (peptidergic). In some cases, the lung sections are also stained for E-cadherin, a marker for airway epithelial cells.
Primary Conclusion: Vagal afferents expressing TRPV1 innervate conducting airways and also project to alveolar region, whereas Tac1 expressing afferent nerves tend to only innervate conducting airways.
Curator's Notes
Experimental Design: AAV9-flex-GFP vectors (RRID:Addgene_51502) were directly injected into anesthetized mice’s sensory ganglia (vagal ganglia and dorsal root ganglia). After four weeks, mice were euthanized and perfused with fixative for lung tissue collection. Mouse lung was then cryosectioned at 80 µm thickness. Lung slices were permeabilized and stained for protein expression using immunohistochemistry. Nonconsecutive entire lung slices were imaged. In many cases, staining of E-Cadherin (RRID:AB_298118), a marker of airway epithelial cell adherens junctions, was used to identify conducting airways from blood vessels. Images were taken with Andor Dragonfly spinning disk confocal microscope equipped with a Zyla 4.2 PLUS sCMOS camera (2048 × 2048 pixels with 6.5-μm pixel size). The pinhole size was 25 μm.
Completeness: This dataset is part of the larger study: Functional mapping of peripheral and central circuits for airway protection and breathing.
Subjects & Samples: Male (n=4) young adult transgenic mice were used in this study.
Primary vs derivative data: Primary data is organized by the subject ID and then sample ID folders. Each folder contains .ims files; Z-stacked multi-tile images stitched using either Fusion software or Imaris Stitcher and further processed using Imaris software. Derivative image data (JPEG2000 and OME-TIFF) was derived from primary images (.ims). Primary images were converted with 20:1 compression to JPEG2000 (.jpx) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Kim, S.-H., Patil, M. J., Hadley, S. H., Bahia, P. K., Butler, S. G., Madaram, M., & Taylor-Clark, T. E. (2022). Mapping of the Sensory Innervation of the Mouse Lung by Specific Vagal and Dorsal Root Ganglion Neuronal Subsets. Eneuro, 9(2), ENEURO.0026-22.2022. https://doi.org/10.1523/eneuro.0026-22.2022
Is Supplemented by
Kim, S.-H. (2022). Dissection and immunohistochemistry of mouse lung v1. https://doi.org/10.17504/protocols.io.3byl4b6kjvo5/v1
Kim, S.-H. (2022). Intraganglionic injection of AAV into nodose ganglia in mice v1. https://doi.org/10.17504/protocols.io.81wgb6w61lpk/v1
