Molecular phenotype distribution of single rat intracardiac neurons
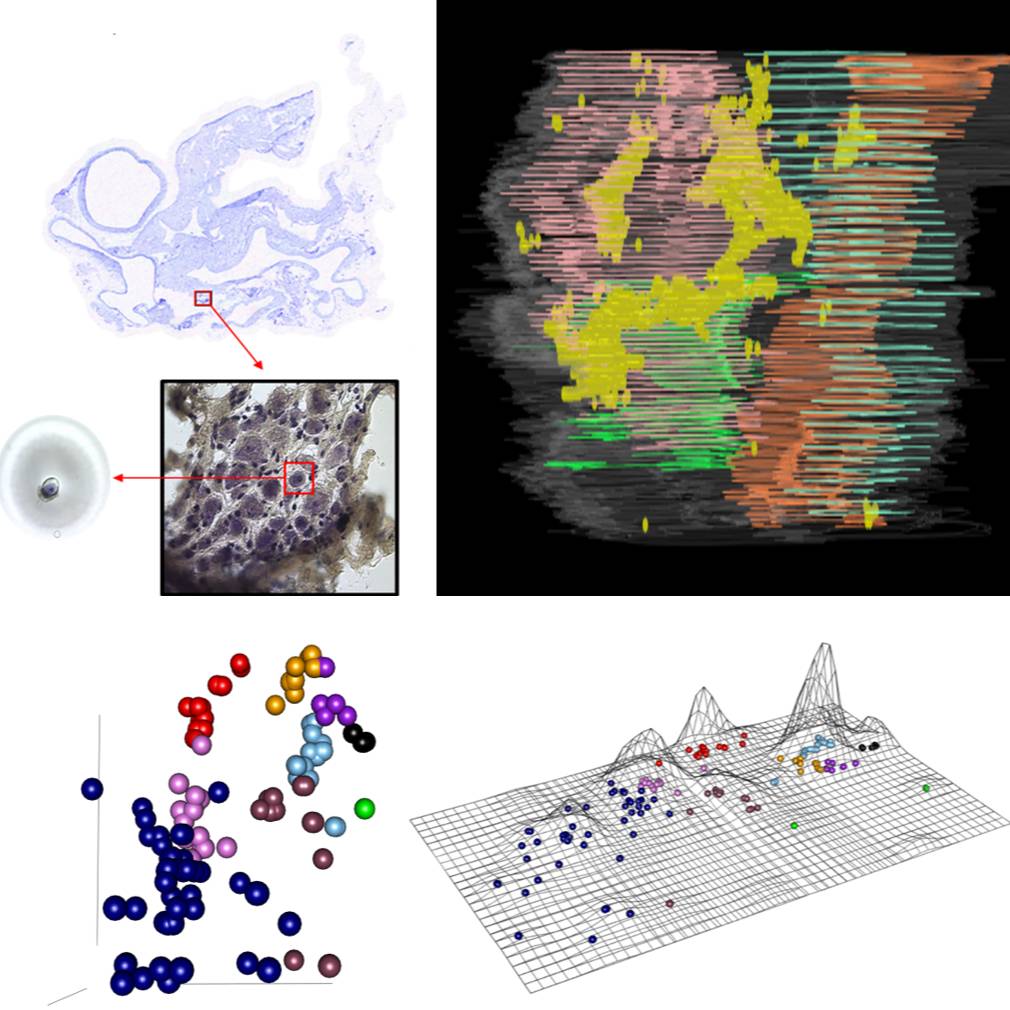
Images collected from serial cryostat sectioning of a cryopreserved heart was used to reconstruct the 3D context. Transcriptional profiles taken from isolated single neurons and mapped back into the previously generated 3D context.
Dataset Overview
Study Purpose: The purpose of this study was to create comprehensive atlas of the cardiac ICN in rat at a cellular level providing gene expression profiles of cardiac neurons on single cell resolution. We developed an approach to appreciate the 3D organization of the intracardiac neurons, ICN, while at the same time permitting single cell transcriptomics and connectomics.
Data Collection: This dataset contains two components. 1. Cardiac neuron gene expression profiling from Real-time quantitative PCR generated by the Fluidigm Dynamic Array: For the data presented in this investigation, data from four 96 well chips were combined into a single matrix and processed further in normalization. The transcriptomic data set consists of 23,254 data points from 151 samples of both single neurons and neuron pools capturing entire clusters. The gene list and primers information available in HB-ICN-4chiprun-design.xlsx file within source folder;. 2. Images of heart samples collected using the Arcturus Laser Capture Microdisection: Images are single sections from a female rat heart in which the 3D organization of the ICN is mapped. The heart was sectioned in the transverse plane going rostro-caudally between the base and apex.
Primary Conclusion: Through serial cryostat sectioning of a cryopreserved heart with imaging of serially collected and stained sections, it is possible to reconstruct the 3D context and collect single neurons using laser microdissection. The transcriptional profiles of these isolated neurons can be determined down to single cell resolution and mapped back into the 3D context generated by stacking the serial images.
Curator's Notes
Experimental Design: The subject was sacrificed using 5% isoflurane inhalation until unconscious and then decapitated. The heart was immediately removed and embedded prior to sectioning, Cresyl Violet staining, and then was imaged. The experimenters used laser capture microdissection to extract single neurons for gene expression analysis.
Completeness: This dataset is part of a larger study: "Mapping of intrinsic cardiac nervous system (ICN) neurons in a 3D reconstructed rat heart."
Subjects & Samples: 10-week-old, female Sprague Dawley rat.
Primary vs derivative data: The primary folder contains sample collection and sample labeling information alongside sample acquisition images. The derivative folder contains normalized data files, spatial mapping data, and supplementary images.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Robbins, S., Moss, A., Nieves, S., & Achanta, S. (2020). Molecular Phenotype Distribution of Single Rat ICN Neurons - Heart B v2. https://doi.org/10.17504/protocols.io.bfxvjpn6
