Mapping colon and bladder innervating sensory neurons in CLARITY cleared ganglia in mouse
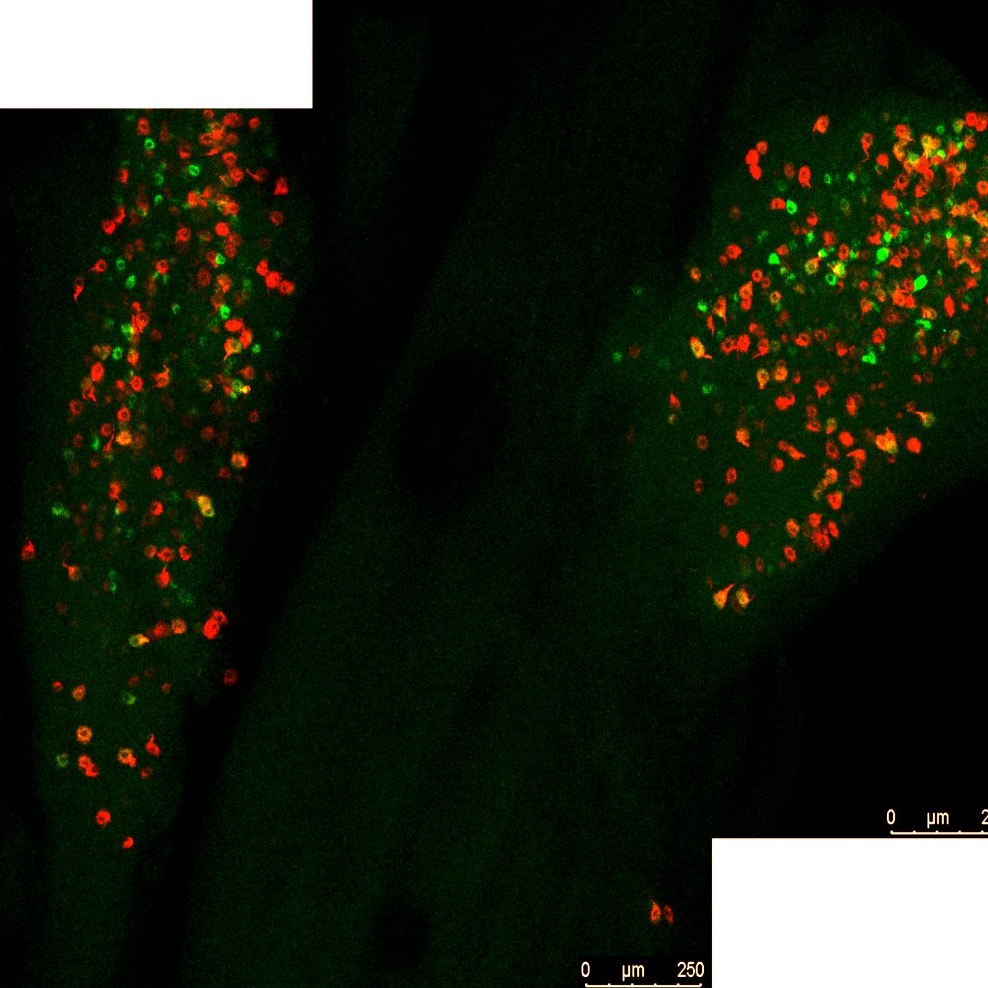
Imaging of colon and bladder retrograde labelled sensory neurons in whole CLARITY cleared dorsal root ganglia and nodose/jugular ganglia complex.
Dataset Overview
Study Purpose: Identify the distribution and proportion of dichotomizing colon/bladder sensory neurons in spinal dorsal root ganglia and vagal nodose/jugular ganglia in the mouse and if this changes in a mouse model of post-colitis chronic visceral hypersensitivity.
Data Collection: Confocal laser scanning microscope (Leica TCS SP8X) was used to optically section cleared ganglia. Confocal images (1024 × 1024 pixels) were obtained with PL APO CS2 air 10X. Sequential scanning (5 line average) was performed with the following settings using a tunable white light laser and photomultiplier detectors: 495 nm excitation and 503 / 538 nm-emission detection for AF488 and 561 nm-excitation and 570-625 nm-emission detection for AF594. Ganglia were optically sectioned (10µm thick), and z-projected images were reconstructed for each ganglia (230-390µm).
Primary Conclusion: Colon and bladder dichotomizing sensory afferent neurons were observed in dorsal root ganglia at spinal levels L5-S1 and T11-L1 as well as in the nodose/jugular ganglia. With the most colon/bladder labeled neurons found within L6 DRG. However, as a proportion of total labeled neurons, those dual labeled were sparse, ranging from 6-10% of the total population. There was no statistical difference in the number or proportion of colon/bladder labeled neurons between sexes, however we saw a reduction in the number and proportion of colon/bladder neurons in L6 DRG in post-colitis mice relative to healthy mice.
Curator's Notes
Experimental Design: Mice underwent retrograde tracing using cholera toxin subunit b (ctb) from the colon (ctb-af594) and bladder (ctb-af488) wall. Four days later, dorsal root ganglia at spinal cord levels thoracic (T9-T13), lumbar (L1-L6), and sacral (S1) were collected, as well as nodose/jugular ganglia complex (N/J). Ganglia underwent clarity clearing, and the ctb labeled neurons were visualized using confocal microscopy in whole ganglia. The number of colon-, bladder- and colon/bladder- labeled neurons were counted per ganglia, and the proportion of each as the total labeled population determined. The number of labelled neurons and the proportions per ganglia was then compared between healthy mice and mice that had a previous bout of colitis. The post-colitis model involved mice receiving an enema of 6.5mg DNBS, and mice that displayed clinical signs of colitis resolving 11 days post DNBS enama underwent retrograde tracing 24 days later. Healthy mice were sex and aged matched to post-colitis mice. N=11 (6m:5f) healthy mice and N=10 (5m:5f) post-colitis mice.
Completeness: This dataset is part of a larger study called "Colon/Blader sensory neurons ganglia in mouse". Related datasets are available.
Subjects & Samples: Male (n=11) and female (n=10) adult mice (RRID:IMSR_JAX:000664) were used in this study.
Primary vs derivative data: Primary data folder contains whole ganglia Z-stacks confocal microscopy LEICA files (.lif). The images are organized in folders by the subject name and sample name, respectively. The number of colonic labeled (red), bladder labeled (green), or dual labeled (yellow) neurons were obtained from projected images of CLARITY cleared ganglia using Image J (NIH) counting tool and is summarized in the excel spreadsheet under derivative data folder. Image data (JPEG2000 and OME-TIFF) was derived from primary images (.lif). Primary images were converted with 40:1 compression to JPEG2000 (.jpx) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal. Primary images were also converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Harrington, A. (2022). Cholera Toxin Subunit B (CTB) Retrograde tracing from the mouse colon and bladder wall. v1. https://doi.org/10.17504/protocols.io.x54v9y391g3e/v1
Harrington, A. (2022). Mapping dichotomisingcolon and bladder sensory afferent neurons and terminals and if they undergo structural plasticity post-colitis. v2. https://doi.org/10.17504/protocols.io.j8nlkk971l5r/v2
Harrington, A. (2022). Mouse model of post-colitis (DNBS) chronic visceral hypersensitivity. v1. https://doi.org/10.17504/protocols.io.14egn7mpqv5d/v1
