Distribution and coexpression patterns of specific cell markers of enteroendocrine cells in pig gastric epithelium
Describes regional differences in gastric wall structure and in the types and densities of hormone secreting cells in the pig stomach.
Dataset Overview
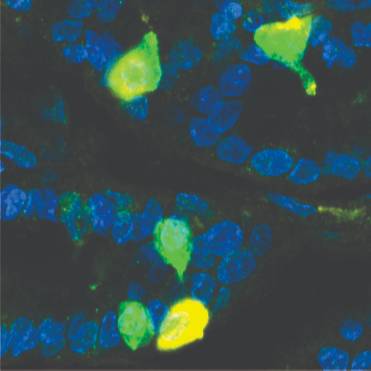
Study Purpose: In this study, we have investigated markers for each of the classes of gastric endocrine cells, gastrin, ghrelin, somatostatin, 5-hydroxytryptamine, and histidine decarboxylase, and PYY cells in pig stomach.
Data Collection: This dataset contains images and analysis used to quantify the number and colocalization of various types of endocrine cells in the pig fundus, corpus, and antrum.
Primary Conclusion: None stated.
Curator's Notes
Experimental Design: In short, pigs were sedated with a xylazil and ketamine mix and euthanised by cardiac injection of pentobarbital sodium (150 mg/kg).Tissues for haematoxylin and eosin staining and immunohistochemistry were removed, opened along the mesenteric border, and pinned flat, mucosa up, without being stretched. Sections for cell counts were imaged as tile scans with a nominal optical thickness of 7.7 μm using a 10x objective on the LSM800 confocal microscope (Zeiss). A 1.5 mm wide region from each imaged section, which contained the full thickness of the mucosa, was selected for analysis in Fiji (http://imagej.nih.gov/ij/). See code>Quantification_macro.ijm and readme.txt file for analysis method.
Completeness: This dataset is a part of a larger study: "Detailed analysis of the intrinsic and extrinsic nerves innervating the pig stomach"
Subjects & Samples: This study used three Large White / Landrace crossbred female pigs (n=4) of unknown age.
Primary vs derivative data: Primary data is organized by subject ID. Sample ID refers to whether tissue is antrum (ant), corpus (cor), or fundus (fun) and which pig it comes from (pig01, pig02, or pig03).RoiSet.zip files contain ROIs for EEC (unnamed ROIs), mucosal areas analysed ("Mucosa", "Basal", "Middle", and "Luminal"), and background regions ("Background"). In derivative data folder Excel "Results_" files contain mean pixel intensity data for all cells in all channels to determine if cell is positive for one or more markers. Sheet a, b, and c contain data for pig01, pig02, and pig03 respectively. The "Results" sheet summarises the thresholded results for each animal, and standardises the data to the total mucosal area, and shows the percentage hormone colocalisation.Data from each "Results_" file is sumarised in the "Quantification_summary.xlsx" file, which is then transfered to the "Quantification_graphs.pzf" graphpad file for producing graphs. For each subject image data (JPEG2000 and OME-TIFF) was derived from primary images (.CZI). .CZI images were converted with 20:1 compression to JPEG2000 (.jpx) by MBF Bioscience for web streaming and visualization on the SPARC Data Portal. .CZI images were also converted with lossless compression to OME-TIFF (.tif) by MBF Bioscience.
Important Notes: Use image J to open the compositive image ("_Comp.tif" file) and the roi file ("_RoiSet.zip") for the selected antibody combination.
Code Availability: Instructions on how to use the code contained in "Quantification_macro.ijm" are described in read me file. Each part of the code (e.g. "PART 1" "PART 2" "PART 3" and "PART 4") can be run independently, by selecting the relevant lines of code and press Control+Shift+R.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Described by
Fothergill, L. J., Galiazzo, G., Hunne, B., Stebbing, M. J., Fakhry, J., Weissenborn, F., Fazio Coles, T. E., & Furness, J. B. (2019). Distribution and co-expression patterns of specific cell markers of enteroendocrine cells in pig gastric epithelium. Cell and Tissue Research, 378(3), 457–469. https://doi.org/10.1007/s00441-019-03065-z
Is Supplemented by
J. Fothergill, L., Stebbing, M., Hunne, B., Galiazzo, G., Fahkry, J., Weissenborn, F., Fazio Coles, T., & B. Furness, J. (2019). Immunohistochemistry and high resolution microscopy of pig gastric enteroendocrine cells v1. https://doi.org/10.17504/protocols.io.4vngw5e
