Intraneural recordings in rat vagus nerves using carbon fiber microelectrode arrays
This data set contains physiological action potential recordings in rat vagus nerves using carbon fiber microelectrode arrays (CFMAs) in spontaneous and modulated conditions.
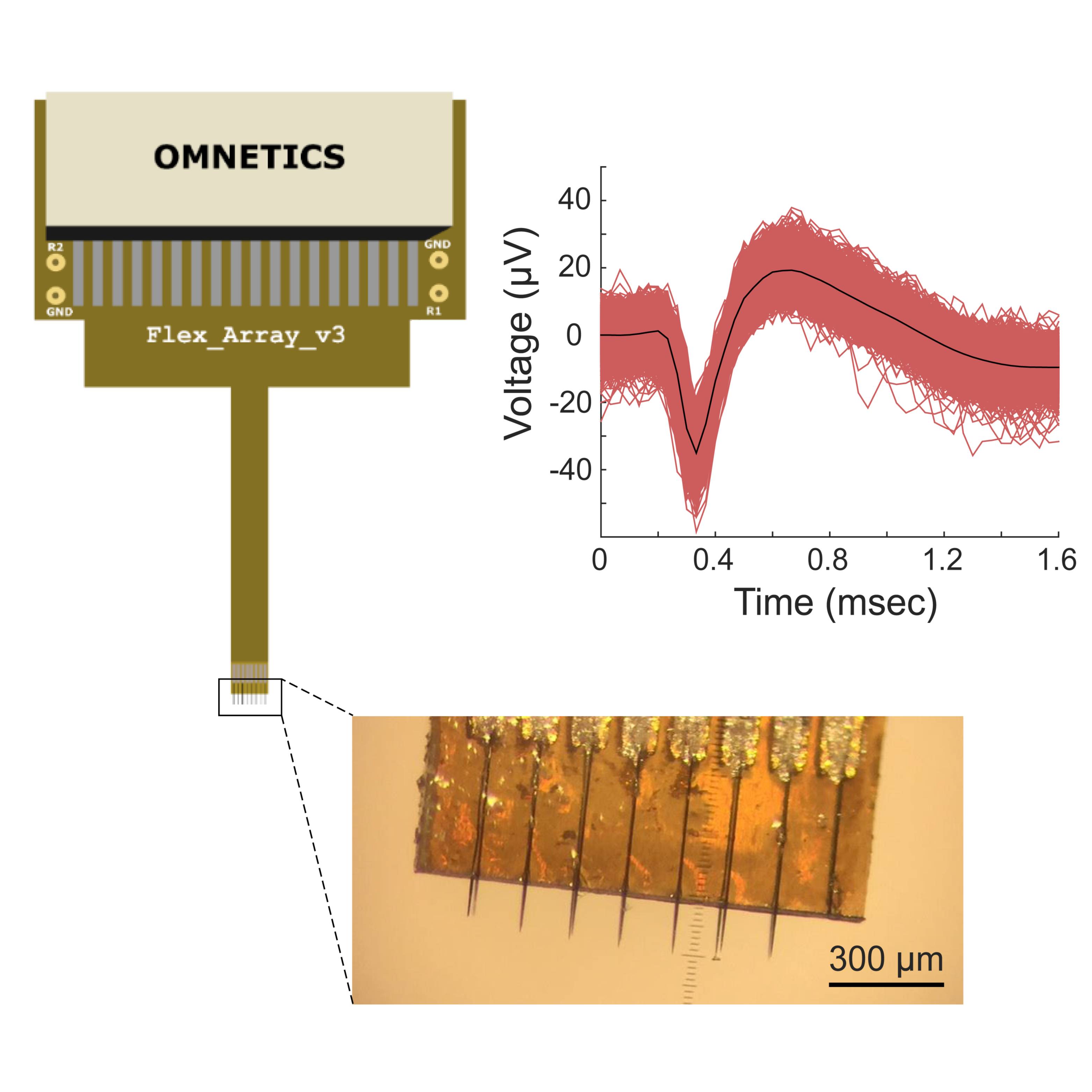
Dataset Overview
Study Purpose: The purpose of this study was to demonstrate physiological intraneural recordings in a small autonomic nerve using the multi-channel carbon fiber microelectrode array (CFMA). This project is part of NIH SPARC award 1OT2OD024907 and NSF award 1707316.
Data Collection: In this study, we inserted CFMA in the cervical vagus nerve of 22 isoflurane-anesthetized rats. The recorded signals were sorted into neural clusters using Wave_Clus and exported to MATLAB for further analysis. We determined the propagation direction and conduction velocity of some vagal signals and quantified the changes in vagal activity in breathing and blood glucose modulated conditions. This data set contains the raw recordings, sorted neural clusters, and analysis codes and outcomes for all the experiments.
Primary Conclusions: Overall, our experiments demonstrated CFMA as a viable multi-channel intraneural electrode for recording neural activity in autonomic nerves. This work provided insights into intraneural autonomic nerve axonal recordings and is a milestone towards the comprehensive understanding of physiological signaling in autonomic nerves.
Curator's Notes
Experimental Design: Using ultra-small carbon fiber microelectrode arrays (CFMAs), physiological action potential recordings from rat vagus nerves were made in the following conditions: spontaneous, blood glucose, and breathing modulated. The rats were anesthetized with isoflurane, which maintained a consistent and stable depth of anesthesia for recording vagal nerve activity. Blood glucose levels were modulated by intraperitoneal (IP) injection of glucose, insulin, or 2-deoxy-D-glucose (2-DG). Breathing was modulated by increasing anesthesia depth.
Completeness: This dataset is complete.
Subjects & Samples: Male (n=19) and female (n=3) Sprague-Dawley rats were used in this study.
Primary vs derivative data: Primary data is organized by the subject ID and contains the converted MATLAB files with waveform and firing rate for all the sorted clusters. The primary data also includes the measured physiological parameters (blood glucose concentration, breathing rate, and heart rate) and impedances. All raw recording files from a Grapevine (Ripple) neural interface processor (.nev and .ns5) are stored in the source folder. The derivative data folder contains the input and output files for spike-sorting of the vagus nerve recordings into neural clusters using Wave_Clus (Chaure et al. 2018, https://doi.org/10.1152/jn.00339.2018).
Code Availability: code folder provides the MATLAB scripts for analyzing the data.
Important Notes: The docs folder contains the input and output image files for spike-sorting the vagus nerve recordings into neural clusters using Wave_Clus (Chaure et al. 2018, https://doi.org/10.1152/jn.00339.2018) arranged by the subject ID. Documents related to the layout and mapping of the CFMA, and the computer-aided design (CAD) for the 3D-printed nerve-holder used in the experiments are also included in the docs folder.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Jiman, A., Ratze, D., Welle, E., Patel, P., Richie, J., Bottorff, E., Seymour, J., Chestek, C., & Bruns, T. (2020). Intraneural Recordings in Rat Vagus Nerves Using Carbon Fiber Microelectrode Arrays v1. https://doi.org/10.17504/protocols.io.bet2jeqe
