Transcriptional diversity of single neurons in the porcine right atrial ganglionic plexus (RAGP)
Single neurons were collected from 4 different porcine RAGP through laser capture microdissection (LCM) and subjected to HT-qPCR to assay for expression of over 200 genes.
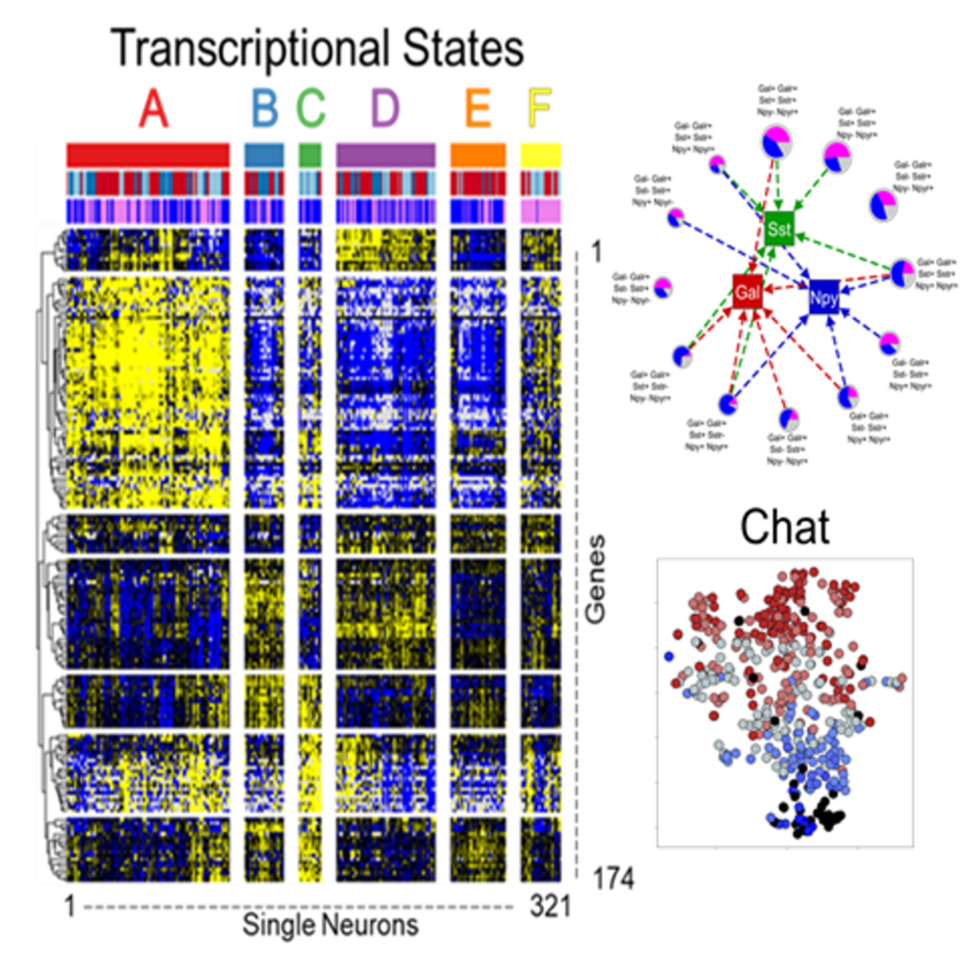
Dataset Overview
Study Purpose: The purpose of this study was to create comprehensive atlas of the cardiac ICN in porcine RAGP at a cellular level providing gene expression profiles of cardiac neurons on single cell resolution. We developed an approach to appreciate the 3D organization of the intracardiac neurons, ICN, while at the same time permitting single cell transcriptomics and connectomics.
Data Collection: The dataset contains neuron gene expression profiling from high throughput real-time quantitative PCR generated by the Fluidigm Dynamic Array: For the data presented in this investigation, data from 21 96-well chips, containing samples from 4 different RAGP were normalized and combined into a single matrix for downstream analysis. The transcriptomic data set consists of 100,015 data points from 241 assayed genes across 415 samples of single neurons across 4 RAGP. The raw output from Fluidigm Biomark is included as well as text files of final matrices showing the Ct values and pass/fail assessment from QC. Data files included are the raw Ct, negative dct, and negative ddct values for 241 genes across 415 samples. A final data matrix, taken forward for analysis, includes 405 samples (10 samples were excluded as outliers) and genes that had more than 40% detection across all samples. Sample annotations are also included.
Primary Conclusion: Through HT-qPCR of single neurons collected across 4 RAGP, it is possible to develop a transcriptional profile of the cardiac ICN in the porcine RAGP. Imaging of these samples as they were collected through Laser Capture Microdissection (LCM) allowed spatial tracking of these samples within their 3D context, allowing the transcriptional profiles of these neurons to be mapped back to their anatomical locations.
Curator's Notes
**Experimental Design:**The porcine heart was injected with FastBlue, a retrograde neuronal tracer, into the SAN before sacrifice, allowing the distinction between SAN-connecting neurons and non SAN-connecting neurons based on whether they stain for just cresyl violet (non SAN-connecting), or cresyl violet and FastBlue (SAN-connecting). Images were taken as an overview of the section as well as at several different levels of magnification both before the samples were lifted and after. Such sample tracking later allowed for mapping of the transcriptomics of the assayed samples back to their 3D locations. Single RAGP neurons were subjected to High-throughput real-time PCR. qRT-PCR reactions were performed using 96.96 BioMark Dynamic Arrays (Fluidigm, South San Francisco, CA, USA) enabling quantitative measurement of multiple mRNAs and samples under identical reaction conditions. Samples from each animal were run across three chips to obtain data on 283 genes per sample. Each chip set runs for a given animal, which contains overlapping assays that serve as technical replicates to evaluate chip-to-chip variability.
Completeness: This dataset is a part of a larger study: Spatially Tracked Single Neuron Anatomical and Molecular Profiling of the Pig Right Atrial Ganglionic Plexus.
Subjects & Samples: Male (n=2) and female (n=2) adolescent Yucatan pigs were used for this study to collect Arcturus Laser Capture Microdissection (LCM), totaling 1215 single-neuron samples.
Primary vs derivative data: There is no primary data in this dataset. Individual qRT-PCR results are included in the source folder. Those include raw results of the qRT-PCR runs across three chips to obtain data on 283 genes per sample. Each set of chip runs for a given animal contained overlapping assays that served as technical replicates to evaluate chip-to-chip variability. The derivative folder contains summarized ddct values, raw Ct values, and sample annotations that include information about which subject the sample was taken from, its connectivity to the SA-Node, section number, and transcriptional state across all 415 samples from all 4 subject RAGP.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Robbins, S., Achanta, S., & Vadigepalli, R. (2021). Laser Capture Microdissection (LCM) and 3D Sample Tracking Protocol. https://doi.org/10.21203/rs.3.pex-927/v1
Achanta, S., & Vadigepalli, R. (2021). Single cell high-throughput qRT-PCR protocol. https://doi.org/10.21203/rs.3.pex-919/v1
Described by
Moss, A., Robbins, S., Achanta, S., Kuttippurathu, L., Turick, S., Nieves, S., Hanna, P., Smith, E. H., Hoover, D. B., Chen, J., Cheng, Z. (Jack), Ardell, J. L., Shivkumar, K., Schwaber, J. S., & Vadigepalli, R. (2021). A single cell transcriptomics map of paracrine networks in the intrinsic cardiac nervous system. IScience, 24(7), 102713. https://doi.org/10.1016/j.isci.2021.102713
