Spatially tracked single-neuron transcriptomics of a male porcine right atrial ganglionic plexus (RAGP)
Single neurons and regional neuronal samples were collected from a male porcine RAGP through laser capture microdissection with preserved spatial tracking.
Dataset Overview
Study Purpose: The purpose of this study was to create comprehensive atlas of the cardiac ICN in rat at a cellular level providing gene expression profiles of cardiac neurons on single cell resolution. We developed an approach to appreciate the 3D organization of the intracardiac neurons, ICN, while at the same time permitting single cell transcriptomics and connectomics.
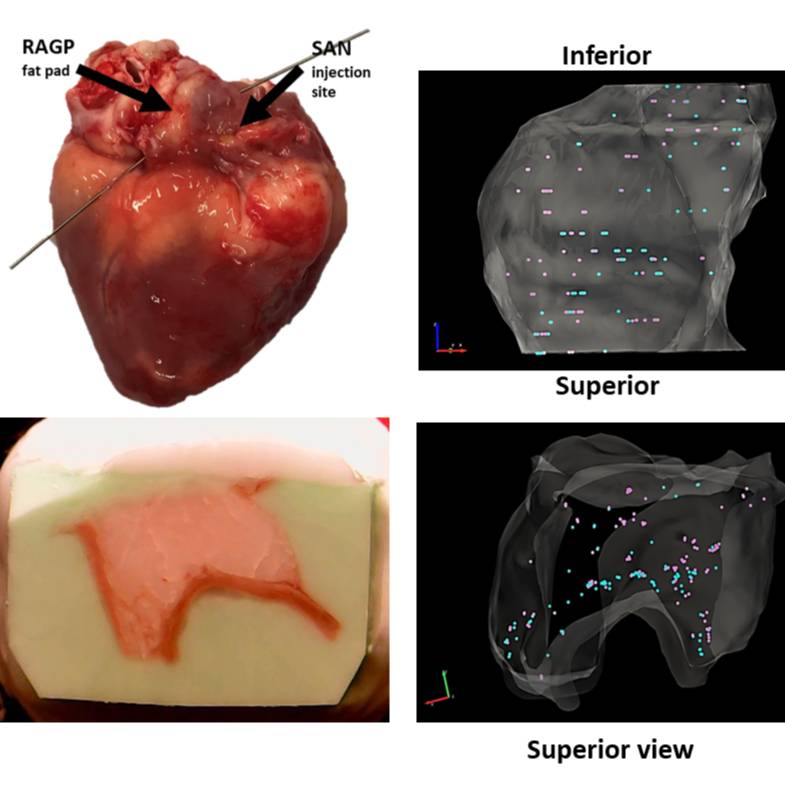
Data Collection: Single neurons subjected to HT-qPCR were collected from a male porcine RAGP. Samples were collected through the use of Laser Capture Microdissection (LCM) allowing for spatial tracking of each sample as it was collected. Through the TissueMaker and TissueMapper software, blockface images taken as the RAGP was sectioned were serially stacked and contoured to provide a wireframe stack of the RAGP. Images taken during LCM sample collection allowed for each sample to be marked within the 3D stack corresponding to its anatomical location. An included JPX file shows the image stack of the RAGP used for reconstruction while an XML files include all contours and marked samples.
Primary Conclusion: Through serial cryostat sectioning of a cryopreserved heart with imaging of serially collected and stained sections, it is possible to reconstruct the 3D context and collect single neurons using LCM. The transcriptional profiles of these isolated neurons can be determined down to single cell resolution and mapped back into the 3D context generated by stacking the serial images.
Curator's Notes
Experimental Design: The RAGP neurons that project to SAN were labeled by a tracer that was injected into the SAN 3 weeks prior to sacrificing the animal for tissue harvesting. The heart tissue corresponding to the location of RAGP was sectioned serially from superior to inferior end for laser capture microdissection of single neurons and neuron pools. During sectioning, blockface images were obtained, which were contoured and organized into a 3D image stack.
Completeness: This dataset is a part of a larger study: Spatially Tracked Single Neuron Anatomical and Molecular Profiling of the Pig Right Atrial Ganglionic Plexus
Subjects & Samples: This study used a single adolescent male Yucatan pig, 43 weeks old.
Primary vs derivative data: Primary data consists of images taken during LCM sample collection that allowed for each sample to be marked within the 3D stack corresponding to its anatomical location. An included JPX file shows the image stack of the RAGP used for reconstruction. The derivative folder contains XML files that include all contours and marked samples, one that shows only RNAseq samples, one only HT-qPCR samples, and one that shows both with RNAseq samples in purple and HT-qPCR samples in yellow.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Robbins, S., Vadigepalli, R., & Schwaber, J. (2021). Single-Cell Mapping and 3D Tissue Reconstruction using Cryosection-derived Images and Tissue Mapper software. https://doi.org/10.21203/rs.3.pex-922/v1
Described by
Moss, A., Robbins, S., Achanta, S., Kuttippurathu, L., Turick, S., Nieves, S., Hanna, P., Smith, E. H., Hoover, D. B., Chen, J., Cheng, Z. (Jack), Ardell, J. L., Shivkumar, K., Schwaber, J. S., & Vadigepalli, R. (2021). A single cell transcriptomics map of paracrine networks in the intrinsic cardiac nervous system. IScience, 24(7), 102713. https://doi.org/10.1016/j.isci.2021.102713
