Spatially tracked single-cell transcriptomics map of neuronal networks in the intrinsic cardiac nervous system
A dataset containing high-resolution figures, supplementary figures, movies and files, as well as the TissueMapper XML annotations and the R code to generate the data-driven plots and visualizations illustrated in Moss et al. 2021
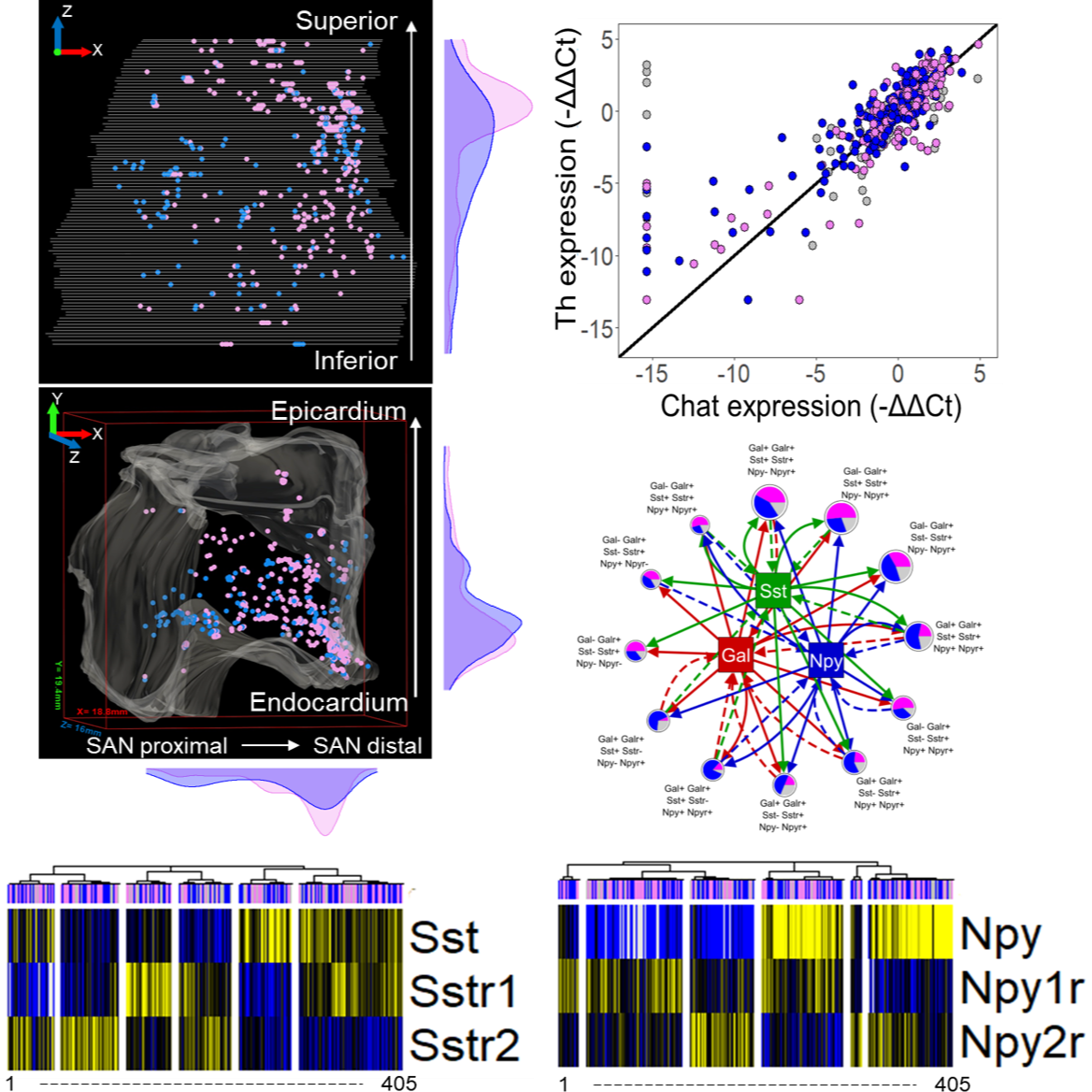
Dataset Overview
Study Purpose: The purpose of this study was to create a comprehensive atlas of the cardiac ICN in porcine Right atrial Ganglionic Plexus (RAGP) at a cellular level providing gene expression profiles of cardiac neurons at single-cell resolution. We developed an approach to appreciate the 3D organization of the intracardiac neurons, ICN, while at the same time permitting single-cell transcriptomics and connectomics.
Data Collection: This dataset contains dataset high-resolution figures, supplementary figures, movies and files, as well as the TissueMapper XML annotations and the R code to generate the data-driven plots and visualizations illustrated in Moss et al. 2021
Primary Conclusion: We developed a spatially-tracked single neuron transcriptomics map of an intrinsic cardiac ganglion - the right atrial ganglionic plexus (RAGP) that is a critical mediator of vagal control of the sinoatrial node (SAN) activity. We developed a 3D representation of RAGP with extensive mapping of neurons and used neuronal tracing to identify the spatial distribution of the subset of neurons that project to the SAN. RNAseq of laser capture microdissected neurons revealed a distinct composition of RAGP neurons compared to CNS neuronal subtypes. High-throughput qPCR of hundreds of laser capture microdissected single neurons led to a surprising finding that cholinergic and catecholaminergic neuronal markers Th and Chat were correlated, suggesting multipotential phenotypes that can drive neuroplasticity within RAGP. Interestingly, no single gene or module was an exclusive marker of RAGP neuronal connectivity to SAN. Neuropeptide-receptor coexpression analysis revealed a combinatorial paracrine neuromodulatory network within RAGP, informing follow-on studies on the vagal control of RAGP to regulate cardiac function in health and disease.
Curator's Notes
**Experimental Design:**The porcine heart was injected with FastBlue, a retrograde neuronal tracer, into the SAN before sacrifice, allowing the distinction between SAN-connecting neurons and non SAN-connecting neurons based on whether they stain for just cresyl violet (non SAN-connecting), or cresyl violet and FastBlue (SAN-connecting). Images were taken of the entire section as well as at several different levels of magnification both before the samples were lifted and after. Such sample tracking later allowed us to map the transcriptomics of the assayed samples back to their 3D locations.
Completeness: This dataset is a part of a larger study: Spatially Tracked Single Neuron Anatomical and Molecular Profiling of the Pig Right Atrial Ganglionic Plexus.
Subjects & Samples: NA (This dataset is based on work with 4 Pigs, however this dataset is the compilation and summary of the associated raw datasets)
Primary vs derivative data: There is no primary data in this dataset. The derivative data consist of summarized annotations for all study samples collected through HT-qPCR, Negddct matrix for 405 samples, and genes that passed QC in at least one RAGP. The Negddct matrix for 415 samples and genes that passed QC in at least one RAGP, and a list of genes that passed QC in all 4 animals are included. The 3D coordinates from all samples processed in PR1643 for 3D visualization are also included in the derivative data folder. The list of genes that had detectable expression across all RAGP and a list of neuronally enriched genes found through GTEx that were present in the RAGP data are also included. Normalized gene counts for single-cell-scale RNAseq of 90 samples from the RAGP.
Files
1 - 0 of 0 files
About this dataset
Publishing history
Cite this dataset
Tags
References
Is Supplemented by
Achanta, S., & Vadigepalli, R. (2021). Single cell high-throughput qRT-PCR protocol. https://doi.org/10.21203/rs.3.pex-919/v1
Robbins, S., Vadigepalli, R., & Schwaber, J. (2021). Single-Cell Mapping and 3D Tissue Reconstruction using Cryosection-derived Images and Tissue Mapper software. https://doi.org/10.21203/rs.3.pex-922/v1
Robbins, S., Achanta, S., & Vadigepalli, R. (2021). Laser Capture Microdissection (LCM) and 3D Sample Tracking Protocol. https://doi.org/10.21203/rs.3.pex-927/v1
Kuttippurathu, L., Moss, A., & Vadigepalli, R. (2021). Single Cell scale RNA-seq Analysis Protocol to analyze Smart-3SEQ data from RAGP neurons of pig heart. https://doi.org/10.21203/rs.3.pex-962/v1
